Vladimir Iglovikov
One Thousand and One Hours: Self-driving Motion Prediction Dataset
Jun 25, 2020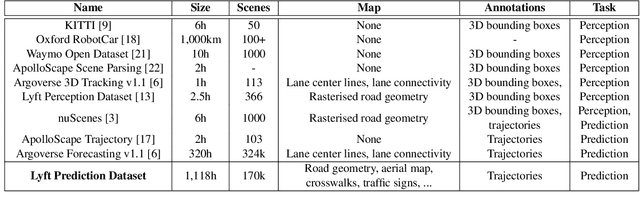
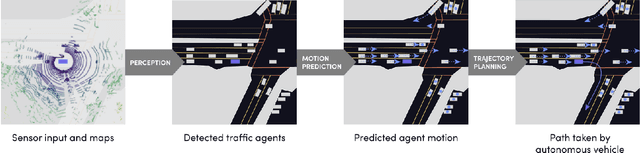
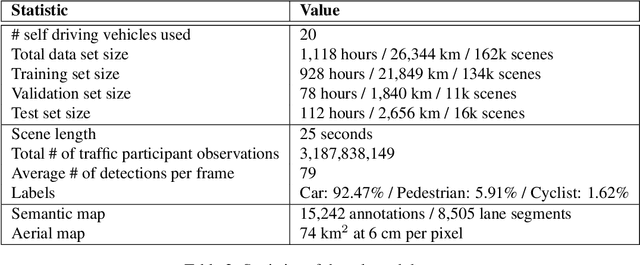
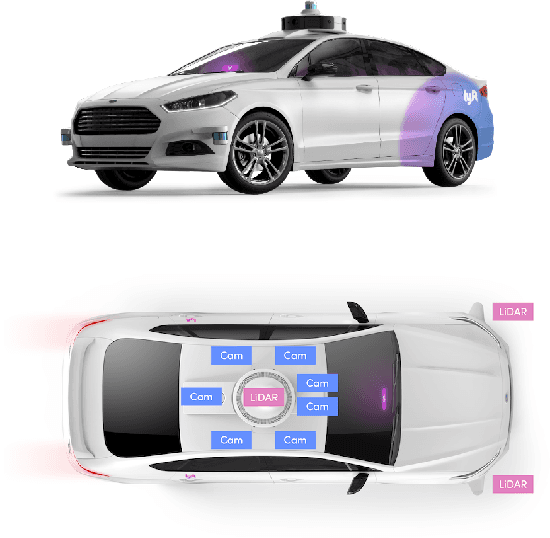
Abstract:We present the largest self-driving dataset for motion prediction to date, with over 1,000 hours of data. This was collected by a fleet of 20 autonomous vehicles along a fixed route in Palo Alto, California over a four-month period. It consists of 170,000 scenes, where each scene is 25 seconds long and captures the perception output of the self-driving system, which encodes the precise positions and motions of nearby vehicles, cyclists, and pedestrians over time. On top of this, the dataset contains a high-definition semantic map with 15,242 labelled elements and a high-definition aerial view over the area. Together with the provided software kit, this collection forms the largest, most complete and detailed dataset to date for the development of self-driving, machine learning tasks such as motion forecasting, planning and simulation. The full dataset is available at http://level5.lyft.com/.
2018 Robotic Scene Segmentation Challenge
Feb 03, 2020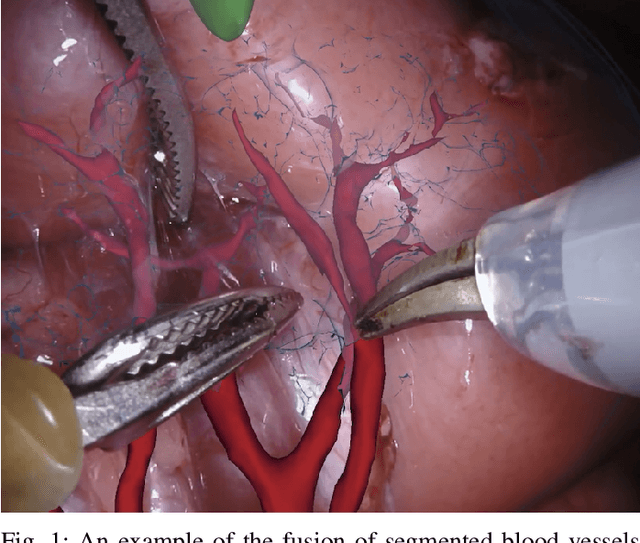
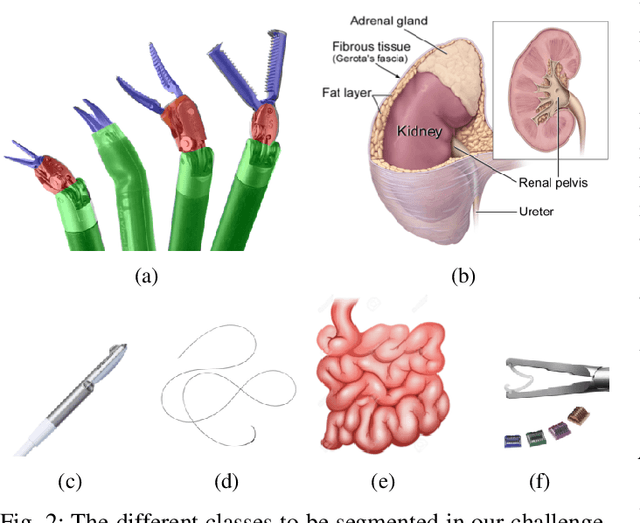
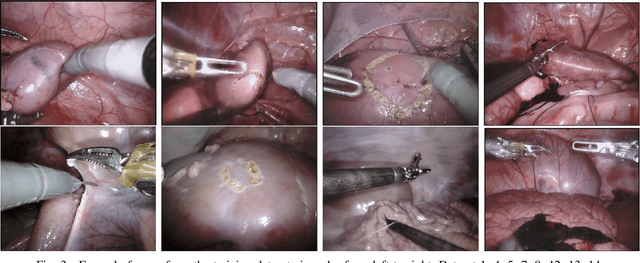
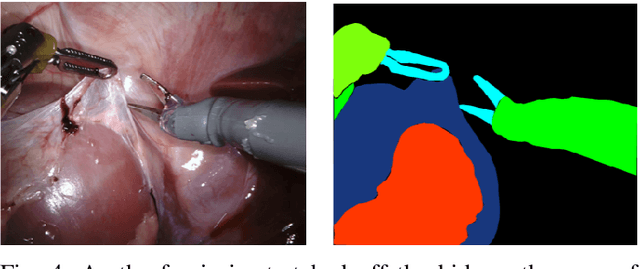
Abstract:In 2015 we began a sub-challenge at the EndoVis workshop at MICCAI in Munich using endoscope images of ex-vivo tissue with automatically generated annotations from robot forward kinematics and instrument CAD models. However, the limited background variation and simple motion rendered the dataset uninformative in learning about which techniques would be suitable for segmentation in real surgery. In 2017, at the same workshop in Quebec we introduced the robotic instrument segmentation dataset with 10 teams participating in the challenge to perform binary, articulating parts and type segmentation of da Vinci instruments. This challenge included realistic instrument motion and more complex porcine tissue as background and was widely addressed with modifications on U-Nets and other popular CNN architectures. In 2018 we added to the complexity by introducing a set of anatomical objects and medical devices to the segmented classes. To avoid over-complicating the challenge, we continued with porcine data which is dramatically simpler than human tissue due to the lack of fatty tissue occluding many organs.
2017 Robotic Instrument Segmentation Challenge
Feb 21, 2019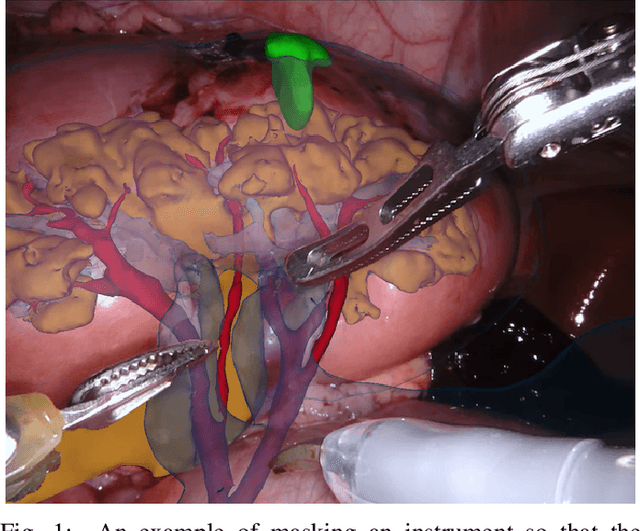
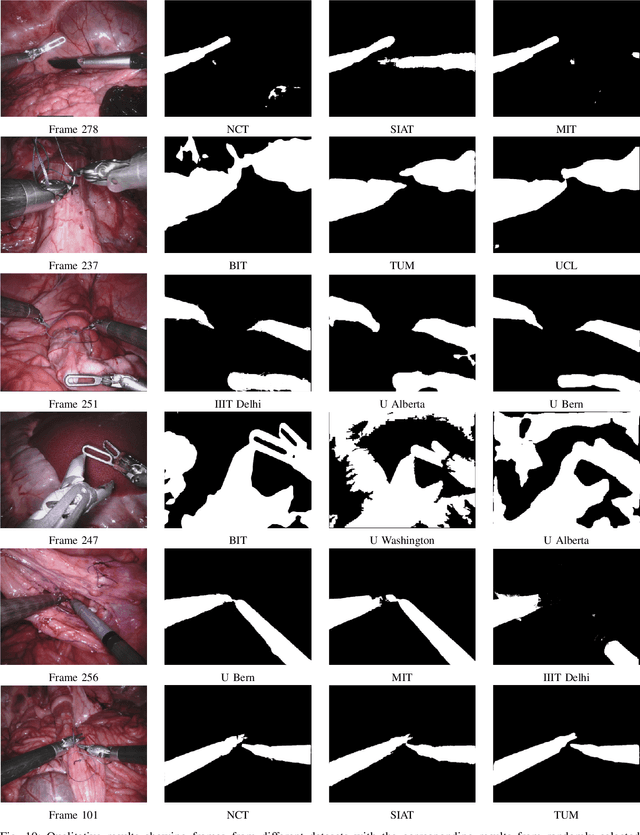
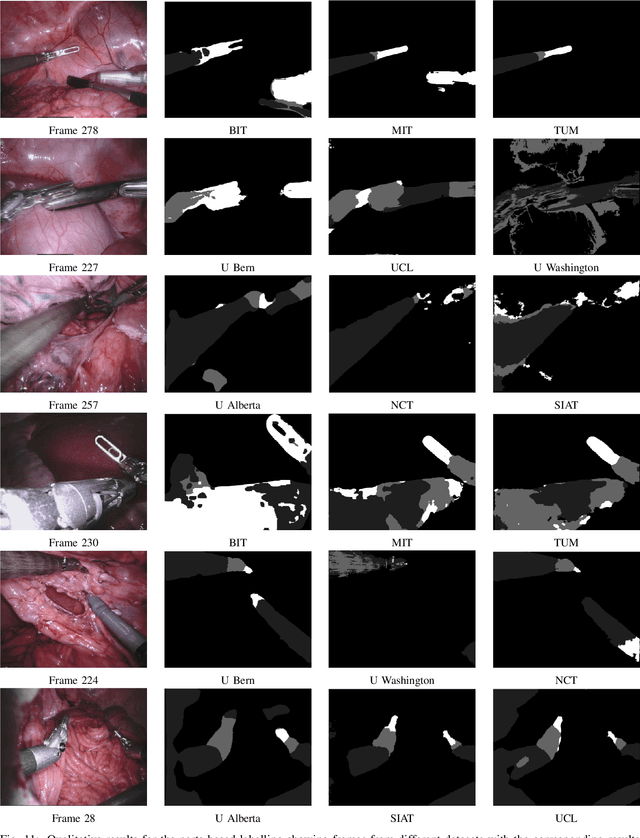

Abstract:In mainstream computer vision and machine learning, public datasets such as ImageNet, COCO and KITTI have helped drive enormous improvements by enabling researchers to understand the strengths and limitations of different algorithms via performance comparison. However, this type of approach has had limited translation to problems in robotic assisted surgery as this field has never established the same level of common datasets and benchmarking methods. In 2015 a sub-challenge was introduced at the EndoVis workshop where a set of robotic images were provided with automatically generated annotations from robot forward kinematics. However, there were issues with this dataset due to the limited background variation, lack of complex motion and inaccuracies in the annotation. In this work we present the results of the 2017 challenge on robotic instrument segmentation which involved 10 teams participating in binary, parts and type based segmentation of articulated da Vinci robotic instruments.
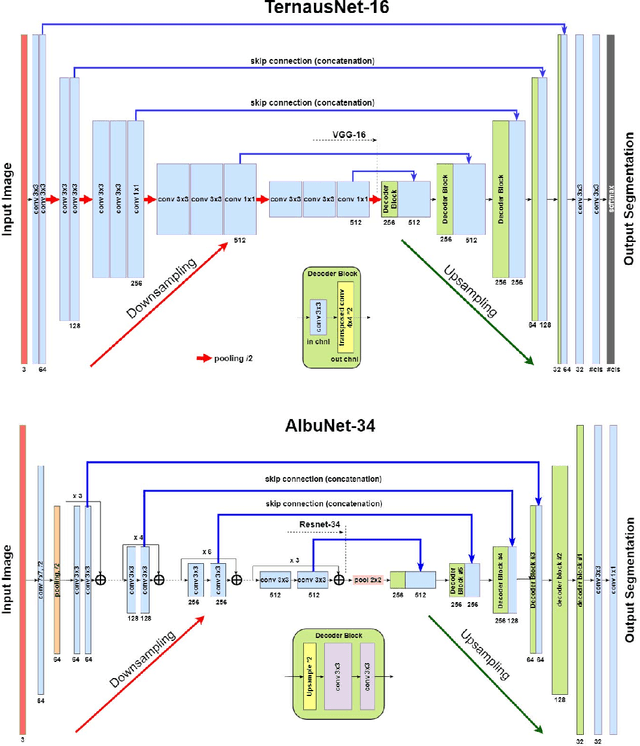
Automatic Instrument Segmentation in Robot-Assisted Surgery Using Deep Learning
Jun 19, 2018
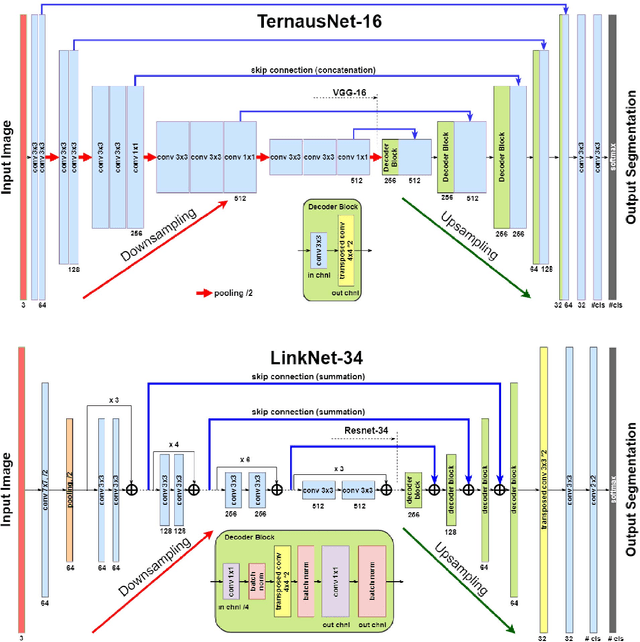
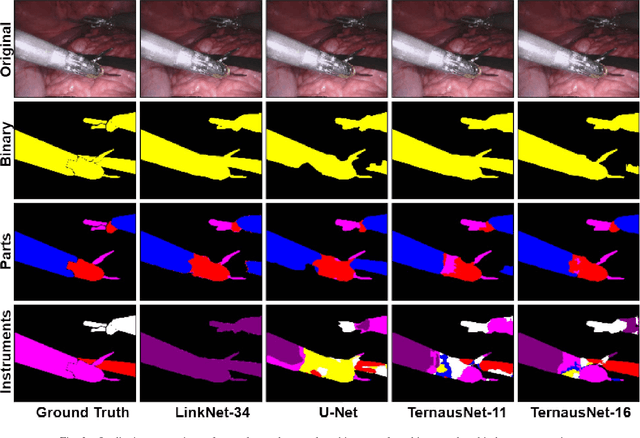
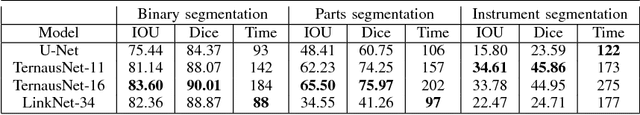
Abstract:Semantic segmentation of robotic instruments is an important problem for the robot-assisted surgery. One of the main challenges is to correctly detect an instrument's position for the tracking and pose estimation in the vicinity of surgical scenes. Accurate pixel-wise instrument segmentation is needed to address this challenge. In this paper we describe our winning solution for MICCAI 2017 Endoscopic Vision SubChallenge: Robotic Instrument Segmentation. Our approach demonstrates an improvement over the state-of-the-art results using several novel deep neural network architectures. It addressed the binary segmentation problem, where every pixel in an image is labeled as an instrument or background from the surgery video feed. In addition, we solve a multi-class segmentation problem, where we distinguish different instruments or different parts of an instrument from the background. In this setting, our approach outperforms other methods in every task subcategory for automatic instrument segmentation thereby providing state-of-the-art solution for this problem. The source code for our solution is made publicly available at https://github.com/ternaus/robot-surgery-segmentation
Pediatric Bone Age Assessment Using Deep Convolutional Neural Networks
Jun 19, 2018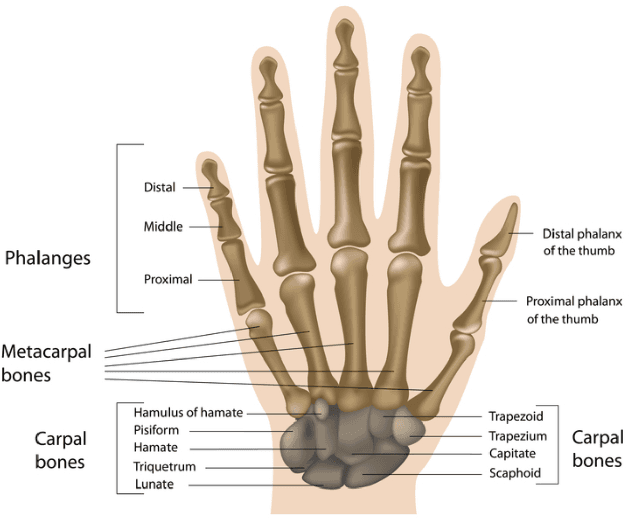

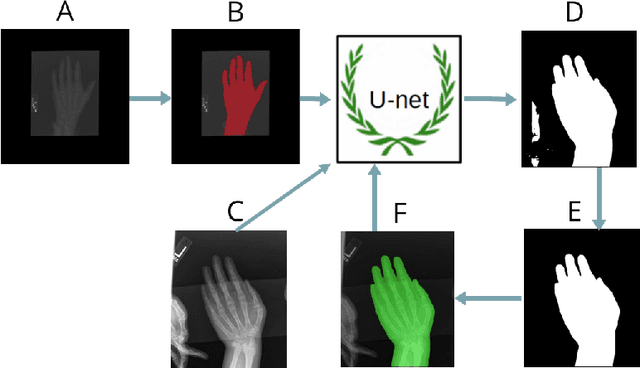
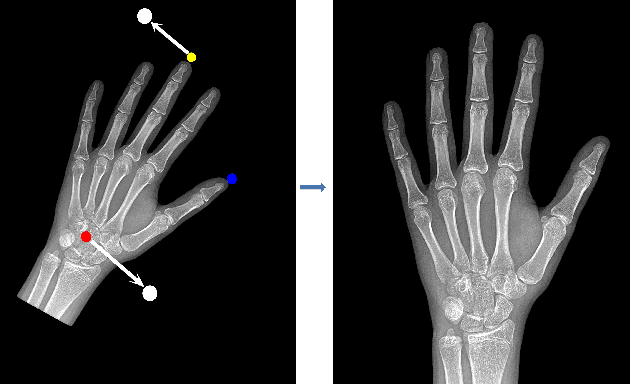
Abstract:Skeletal bone age assessment is a common clinical practice to diagnose endocrine and metabolic disorders in child development. In this paper, we describe a fully automated deep learning approach to the problem of bone age assessment using data from Pediatric Bone Age Challenge organized by RSNA 2017. The dataset for this competition is consisted of 12.6k radiological images of left hand labeled by the bone age and sex of patients. Our approach utilizes several deep learning architectures: U-Net, ResNet-50, and custom VGG-style neural networks trained end-to-end. We use images of whole hands as well as specific parts of a hand for both training and inference. This approach allows us to measure importance of specific hand bones for the automated bone age analysis. We further evaluate performance of the method in the context of skeletal development stages. Our approach outperforms other common methods for bone age assessment.
Angiodysplasia Detection and Localization Using Deep Convolutional Neural Networks
Apr 21, 2018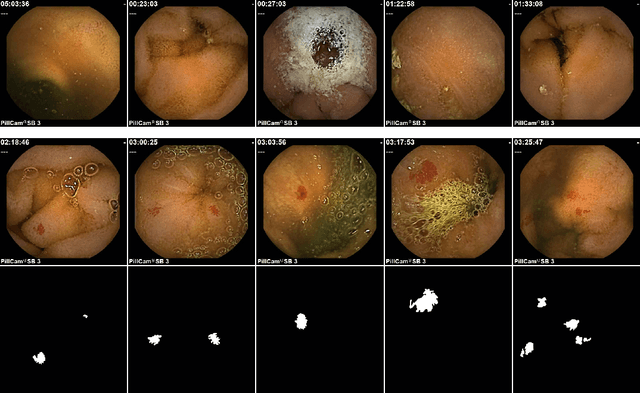

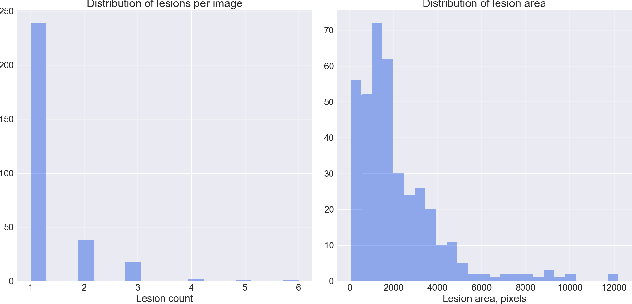
Abstract:Accurate detection and localization for angiodysplasia lesions is an important problem in early stage diagnostics of gastrointestinal bleeding and anemia. Gold-standard for angiodysplasia detection and localization is performed using wireless capsule endoscopy. This pill-like device is able to produce thousand of high enough resolution images during one passage through gastrointestinal tract. In this paper we present our winning solution for MICCAI 2017 Endoscopic Vision SubChallenge: Angiodysplasia Detection and Localization its further improvements over the state-of-the-art results using several novel deep neural network architectures. It address the binary segmentation problem, where every pixel in an image is labeled as an angiodysplasia lesions or background. Then, we analyze connected component of each predicted mask. Based on the analysis we developed a classifier that predict angiodysplasia lesions (binary variable) and a detector for their localization (center of a component). In this setting, our approach outperforms other methods in every task subcategory for angiodysplasia detection and localization thereby providing state-of-the-art results for these problems. The source code for our solution is made publicly available at https://github.com/ternaus/angiodysplasia-segmentatio
Deep Convolutional Neural Networks for Breast Cancer Histology Image Analysis
Apr 03, 2018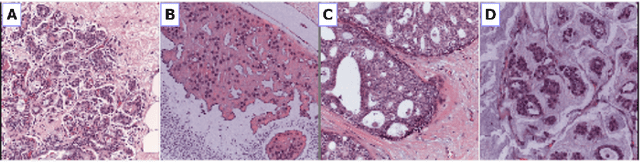
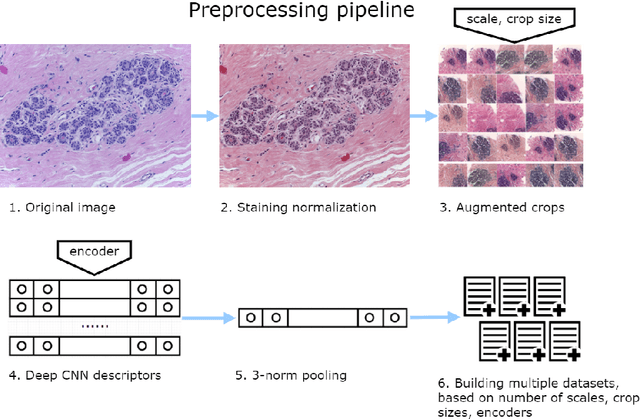
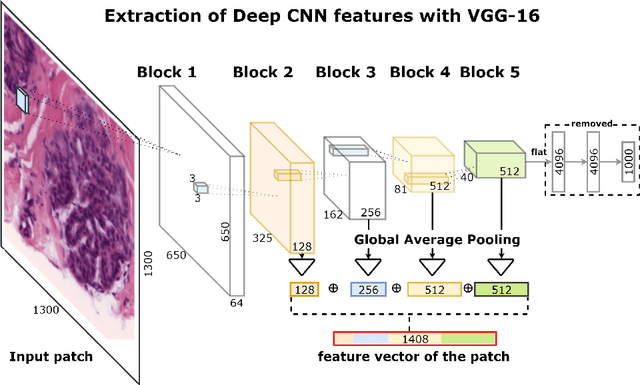
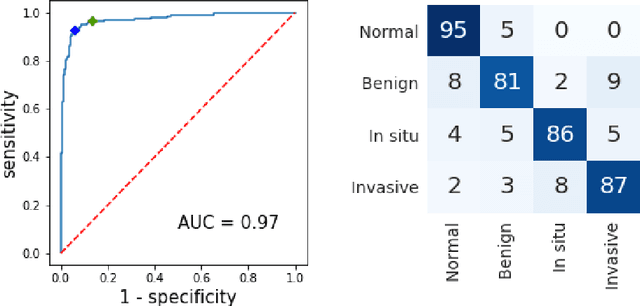
Abstract:Breast cancer is one of the main causes of cancer death worldwide. Early diagnostics significantly increases the chances of correct treatment and survival, but this process is tedious and often leads to a disagreement between pathologists. Computer-aided diagnosis systems showed potential for improving the diagnostic accuracy. In this work, we develop the computational approach based on deep convolution neural networks for breast cancer histology image classification. Hematoxylin and eosin stained breast histology microscopy image dataset is provided as a part of the ICIAR 2018 Grand Challenge on Breast Cancer Histology Images. Our approach utilizes several deep neural network architectures and gradient boosted trees classifier. For 4-class classification task, we report 87.2% accuracy. For 2-class classification task to detect carcinomas we report 93.8% accuracy, AUC 97.3%, and sensitivity/specificity 96.5/88.0% at the high-sensitivity operating point. To our knowledge, this approach outperforms other common methods in automated histopathological image classification. The source code for our approach is made publicly available at https://github.com/alexander-rakhlin/ICIAR2018
TernausNet: U-Net with VGG11 Encoder Pre-Trained on ImageNet for Image Segmentation
Jan 17, 2018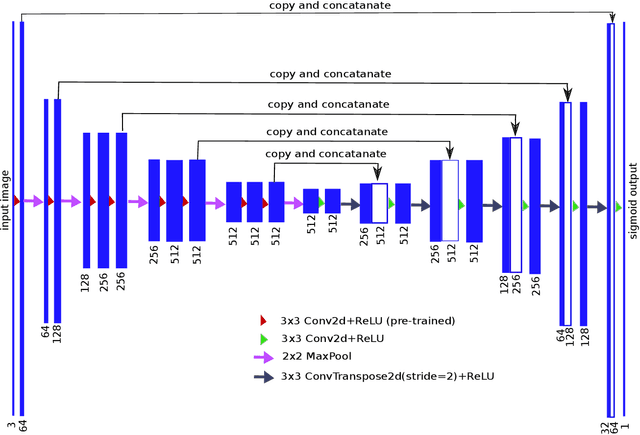
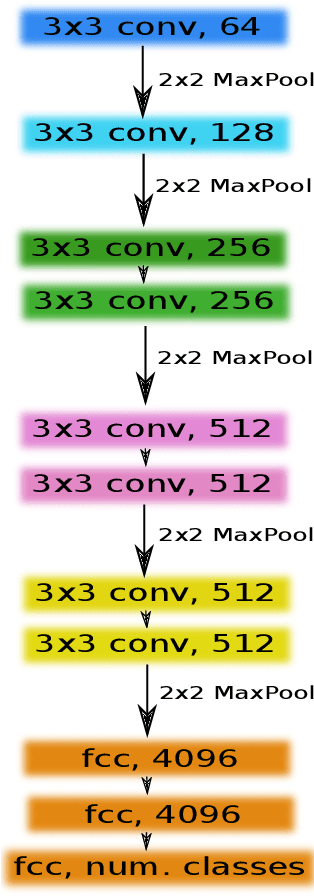
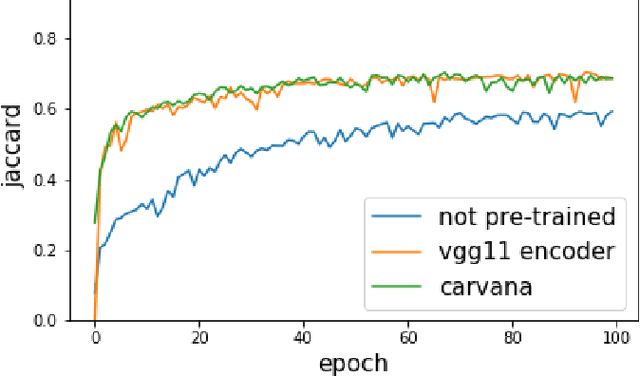

Abstract:Pixel-wise image segmentation is demanding task in computer vision. Classical U-Net architectures composed of encoders and decoders are very popular for segmentation of medical images, satellite images etc. Typically, neural network initialized with weights from a network pre-trained on a large data set like ImageNet shows better performance than those trained from scratch on a small dataset. In some practical applications, particularly in medicine and traffic safety, the accuracy of the models is of utmost importance. In this paper, we demonstrate how the U-Net type architecture can be improved by the use of the pre-trained encoder. Our code and corresponding pre-trained weights are publicly available at https://github.com/ternaus/TernausNet. We compare three weight initialization schemes: LeCun uniform, the encoder with weights from VGG11 and full network trained on the Carvana dataset. This network architecture was a part of the winning solution (1st out of 735) in the Kaggle: Carvana Image Masking Challenge.
Satellite Imagery Feature Detection using Deep Convolutional Neural Network: A Kaggle Competition
Jun 19, 2017
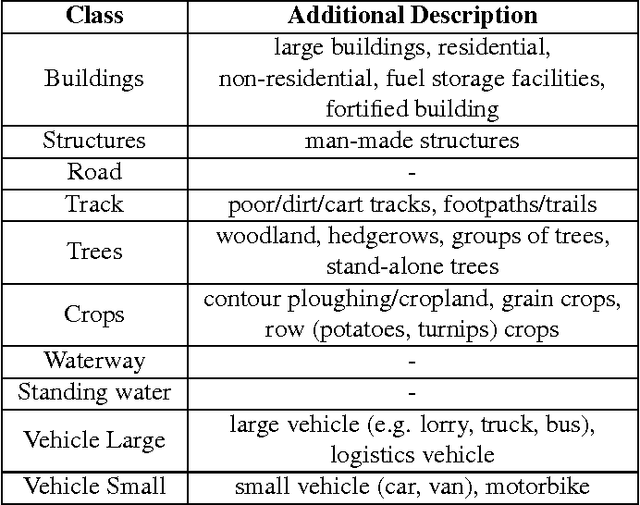
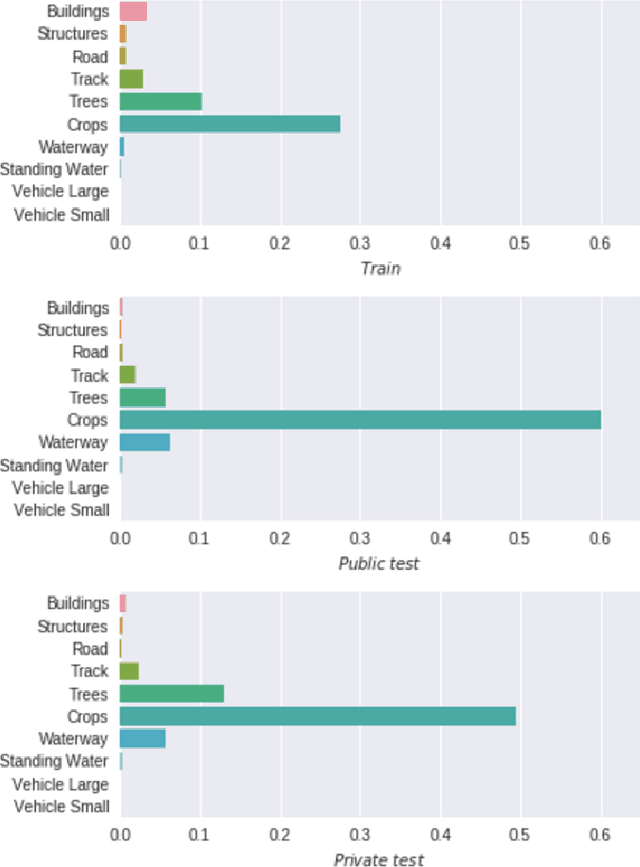
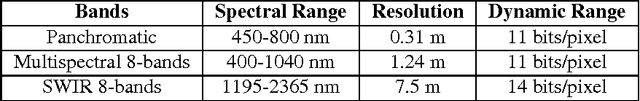
Abstract:This paper describes our approach to the DSTL Satellite Imagery Feature Detection challenge run by Kaggle. The primary goal of this challenge is accurate semantic segmentation of different classes in satellite imagery. Our approach is based on an adaptation of fully convolutional neural network for multispectral data processing. In addition, we defined several modifications to the training objective and overall training pipeline, e.g. boundary effect estimation, also we discuss usage of data augmentation strategies and reflectance indices. Our solution scored third place out of 419 entries. Its accuracy is comparable to the first two places, but unlike those solutions, it doesn't rely on complex ensembling techniques and thus can be easily scaled for deployment in production as a part of automatic feature labeling systems for satellite imagery analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge