Ahmed Mohammed
Multi-label Video Classification for Underwater Ship Inspection
May 27, 2023



Abstract:Today ship hull inspection including the examination of the external coating, detection of defects, and other types of external degradation such as corrosion and marine growth is conducted underwater by means of Remotely Operated Vehicles (ROVs). The inspection process consists of a manual video analysis which is a time-consuming and labor-intensive process. To address this, we propose an automatic video analysis system using deep learning and computer vision to improve upon existing methods that only consider spatial information on individual frames in underwater ship hull video inspection. By exploring the benefits of adding temporal information and analyzing frame-based classifiers, we propose a multi-label video classification model that exploits the self-attention mechanism of transformers to capture spatiotemporal attention in consecutive video frames. Our proposed method has demonstrated promising results and can serve as a benchmark for future research and development in underwater video inspection applications.
Evaluating clinical diversity and plausibility of synthetic capsule endoscopic images
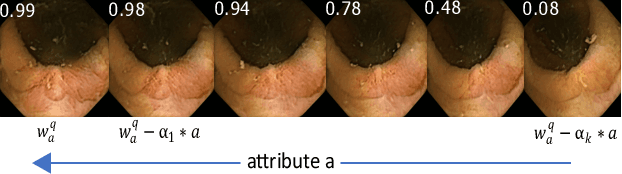
Jan 16, 2023Abstract:Wireless Capsule Endoscopy (WCE) is being increasingly used as an alternative imaging modality for complete and non-invasive screening of the gastrointestinal tract. Although this is advantageous in reducing unnecessary hospital admissions, it also demands that a WCE diagnostic protocol be in place so larger populations can be effectively screened. This calls for training and education protocols attuned specifically to this modality. Like training in other modalities such as traditional endoscopy, CT, MRI, etc., a WCE training protocol would require an atlas comprising of a large corpora of images that show vivid descriptions of pathologies and abnormalities, ideally observed over a period of time. Since such comprehensive atlases are presently lacking in WCE, in this work, we propose a deep learning method for utilizing already available studies across different institutions for the creation of a realistic WCE atlas using StyleGAN. We identify clinically relevant attributes in WCE such that synthetic images can be generated with selected attributes on cue. Beyond this, we also simulate several disease progression scenarios. The generated images are evaluated for realism and plausibility through three subjective online experiments with the participation of eight gastroenterology experts from three geographical locations and a variety of years of experience. The results from the experiments indicate that the images are highly realistic and the disease scenarios plausible. The images comprising the atlas are available publicly for use in training applications as well as supplementing real datasets for deep learning.
This changes to that : Combining causal and non-causal explanations to generate disease progression in capsule endoscopy
Dec 05, 2022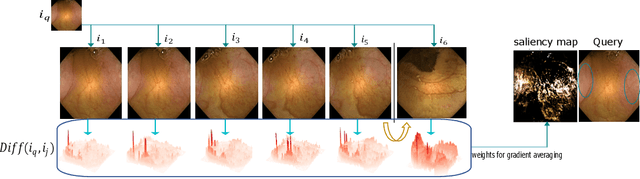
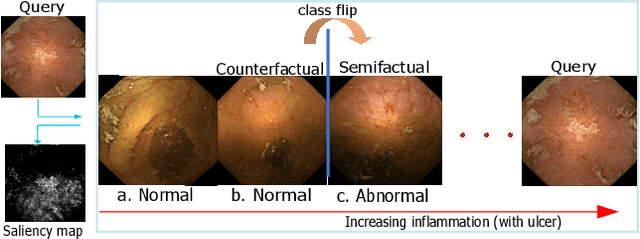
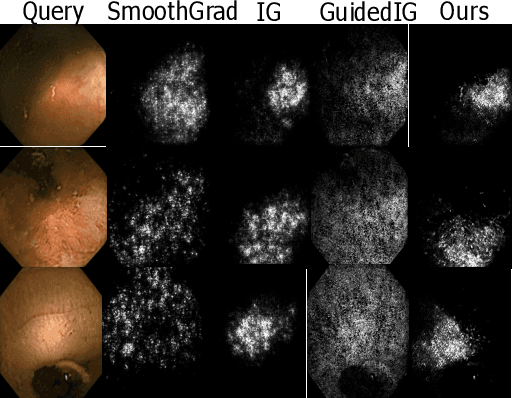
Abstract:Due to the unequivocal need for understanding the decision processes of deep learning networks, both modal-dependent and model-agnostic techniques have become very popular. Although both of these ideas provide transparency for automated decision making, most methodologies focus on either using the modal-gradients (model-dependent) or ignoring the model internal states and reasoning with a model's behavior/outcome (model-agnostic) to instances. In this work, we propose a unified explanation approach that given an instance combines both model-dependent and agnostic explanations to produce an explanation set. The generated explanations are not only consistent in the neighborhood of a sample but can highlight causal relationships between image content and the outcome. We use Wireless Capsule Endoscopy (WCE) domain to illustrate the effectiveness of our explanations. The saliency maps generated by our approach are comparable or better on the softmax information score.
From Labels to Priors in Capsule Endoscopy: A Prior Guided Approach for Improving Generalization with Few Labels
Jun 10, 2022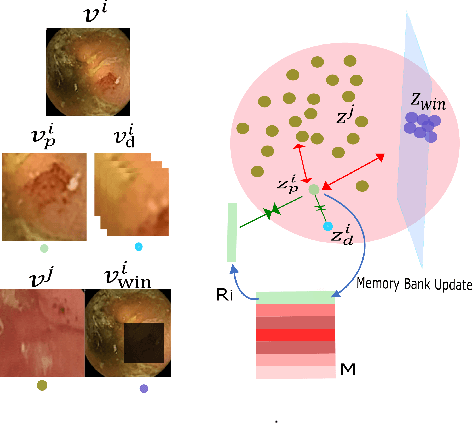

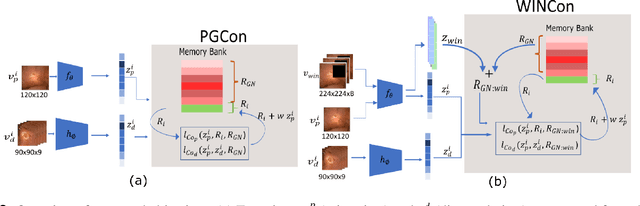
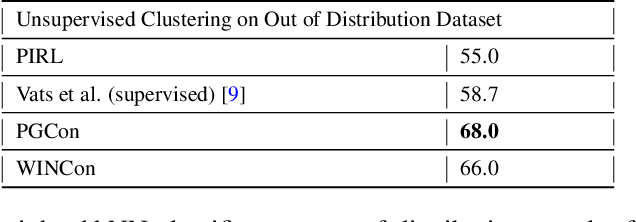
Abstract:The lack of generalizability of deep learning approaches for the automated diagnosis of pathologies in Wireless Capsule Endoscopy (WCE) has prevented any significant advantages from trickling down to real clinical practices. As a result, disease management using WCE continues to depend on exhaustive manual investigations by medical experts. This explains its limited use despite several advantages. Prior works have considered using higher quality and quantity of labels as a way of tackling the lack of generalization, however this is hardly scalable considering pathology diversity not to mention that labeling large datasets encumbers the medical staff additionally. We propose using freely available domain knowledge as priors to learn more robust and generalizable representations. We experimentally show that domain priors can benefit representations by acting in proxy of labels, thereby significantly reducing the labeling requirement while still enabling fully unsupervised yet pathology-aware learning. We use the contrastive objective along with prior-guided views during pretraining, where the view choices inspire sensitivity to pathological information. Extensive experiments on three datasets show that our method performs better than (or closes gap with) the state-of-the-art in the domain, establishing a new benchmark in pathology classification and cross-dataset generalization, as well as scaling to unseen pathology categories.
Learning More for Free - A Multi Task Learning Approach for Improved Pathology Classification in Capsule Endoscopy
Jun 30, 2021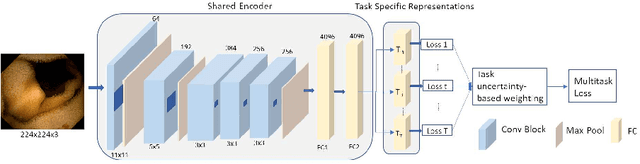
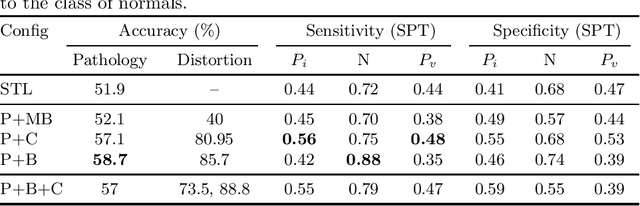
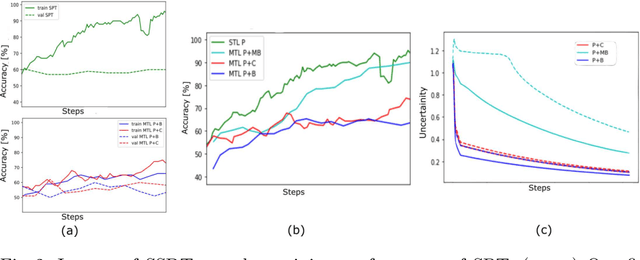

Abstract:The progress in Computer Aided Diagnosis (CADx) of Wireless Capsule Endoscopy (WCE) is thwarted by the lack of data. The inadequacy in richly representative healthy and abnormal conditions results in isolated analyses of pathologies, that can not handle realistic multi-pathology scenarios. In this work, we explore how to learn more for free, from limited data through solving a WCE multicentric, multi-pathology classification problem. Learning more implies to learning more than full supervision would allow with the same data. This is done by combining self supervision with full supervision, under multi task learning. Additionally, we draw inspiration from the Human Visual System (HVS) in designing self supervision tasks and investigate if seemingly ineffectual signals within the data itself can be exploited to gain performance, if so, which signals would be better than others. Further, we present our analysis of the high level features as a stepping stone towards more robust multi-pathology CADx in WCE.
2018 Robotic Scene Segmentation Challenge
Feb 03, 2020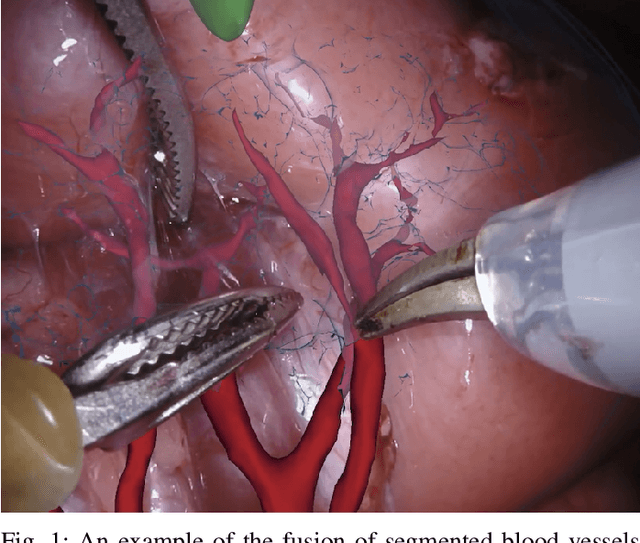
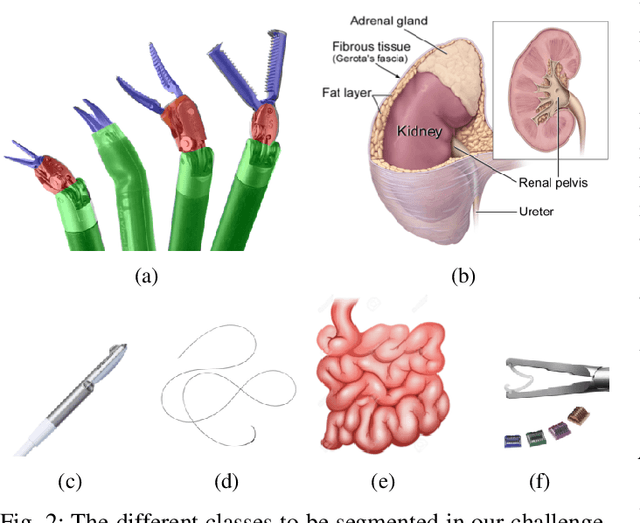
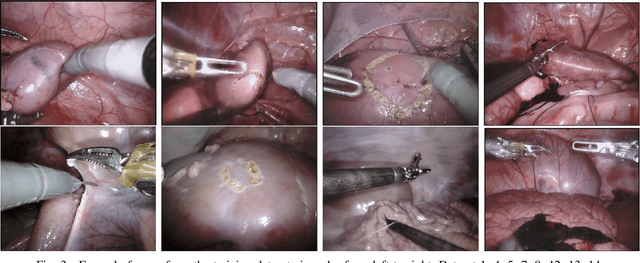
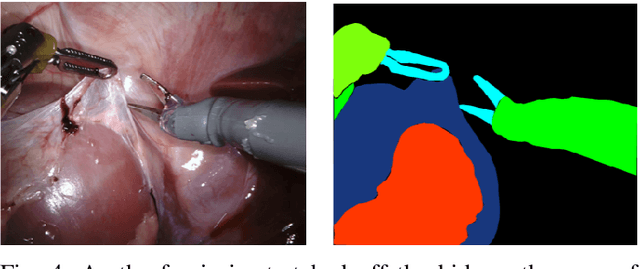
Abstract:In 2015 we began a sub-challenge at the EndoVis workshop at MICCAI in Munich using endoscope images of ex-vivo tissue with automatically generated annotations from robot forward kinematics and instrument CAD models. However, the limited background variation and simple motion rendered the dataset uninformative in learning about which techniques would be suitable for segmentation in real surgery. In 2017, at the same workshop in Quebec we introduced the robotic instrument segmentation dataset with 10 teams participating in the challenge to perform binary, articulating parts and type segmentation of da Vinci instruments. This challenge included realistic instrument motion and more complex porcine tissue as background and was widely addressed with modifications on U-Nets and other popular CNN architectures. In 2018 we added to the complexity by introducing a set of anatomical objects and medical devices to the segmented classes. To avoid over-complicating the challenge, we continued with porcine data which is dramatically simpler than human tissue due to the lack of fatty tissue occluding many organs.
Y-Net: A deep Convolutional Neural Network for Polyp Detection
Jun 05, 2018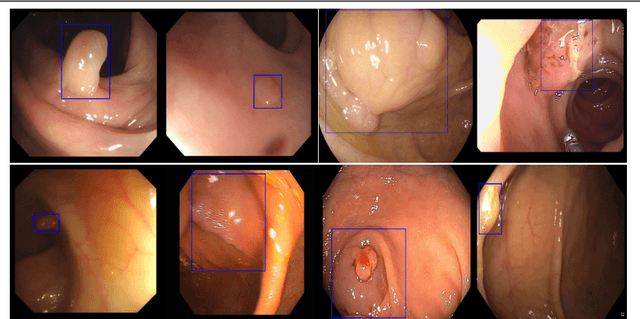
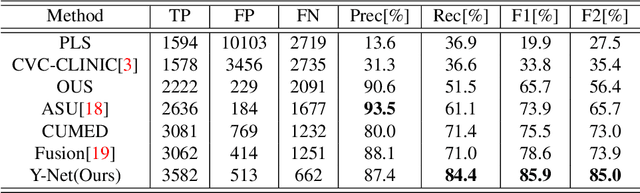
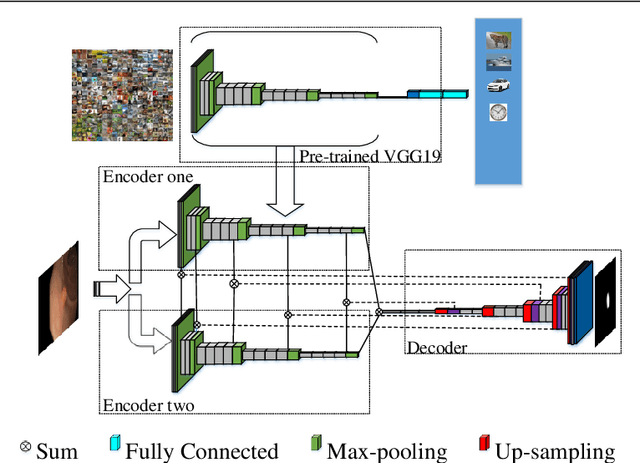

Abstract:Colorectal polyps are important precursors to colon cancer, the third most common cause of cancer mortality for both men and women. It is a disease where early detection is of crucial importance. Colonoscopy is commonly used for early detection of cancer and precancerous pathology. It is a demanding procedure requiring significant amount of time from specialized physicians and nurses, in addition to a significant miss-rates of polyps by specialists. Automated polyp detection in colonoscopy videos has been demonstrated to be a promising way to handle this problem. {However, polyps detection is a challenging problem due to the availability of limited amount of training data and large appearance variations of polyps. To handle this problem, we propose a novel deep learning method Y-Net that consists of two encoder networks with a decoder network. Our proposed Y-Net method} relies on efficient use of pre-trained and un-trained models with novel sum-skip-concatenation operations. Each of the encoders are trained with encoder specific learning rate along the decoder. Compared with the previous methods employing hand-crafted features or 2-D/3-D convolutional neural network, our approach outperforms state-of-the-art methods for polyp detection with 7.3% F1-score and 13% recall improvement.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge