Stefan Leger
Graph data modelling for outcome prediction in oropharyngeal cancer patients
Oct 04, 2023



Abstract:Graph neural networks (GNNs) are becoming increasingly popular in the medical domain for the tasks of disease classification and outcome prediction. Since patient data is not readily available as a graph, most existing methods either manually define a patient graph, or learn a latent graph based on pairwise similarities between the patients. There are also hypergraph neural network (HGNN)-based methods that were introduced recently to exploit potential higher order associations between the patients by representing them as a hypergraph. In this work, we propose a patient hypergraph network (PHGN), which has been investigated in an inductive learning setup for binary outcome prediction in oropharyngeal cancer (OPC) patients using computed tomography (CT)-based radiomic features for the first time. Additionally, the proposed model was extended to perform time-to-event analyses, and compared with GNN and baseline linear models.
Non-Rigid Volume to Surface Registration using a Data-Driven Biomechanical Model
May 29, 2020



Abstract:Non-rigid registration is a key component in soft-tissue navigation. We focus on laparoscopic liver surgery, where we register the organ model obtained from a preoperative CT scan to the intraoperative partial organ surface, reconstructed from the laparoscopic video. This is a challenging task due to sparse and noisy intraoperative data, real-time requirements and many unknowns - such as tissue properties and boundary conditions. Furthermore, establishing correspondences between pre- and intraoperative data can be extremely difficult since the liver usually lacks distinct surface features and the used imaging modalities suffer from very different types of noise. In this work, we train a convolutional neural network to perform both the search for surface correspondences as well as the non-rigid registration in one step. The network is trained on physically accurate biomechanical simulations of randomly generated, deforming organ-like structures. This enables the network to immediately generalize to a new patient organ without the need to re-train. We add various amounts of noise to the intraoperative surfaces during training, making the network robust to noisy intraoperative data. During inference, the network outputs the displacement field which matches the preoperative volume to the partial intraoperative surface. In multiple experiments, we show that the network translates well to real data while maintaining a high inference speed. Our code is made available online.
Robust Medical Instrument Segmentation Challenge 2019
Mar 23, 2020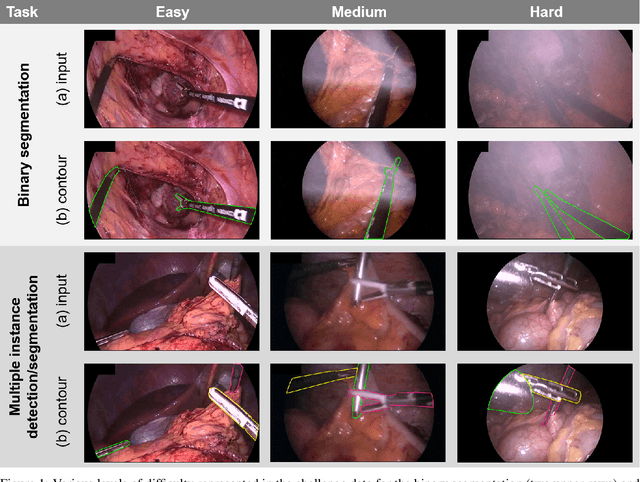
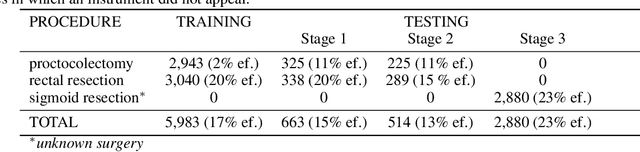
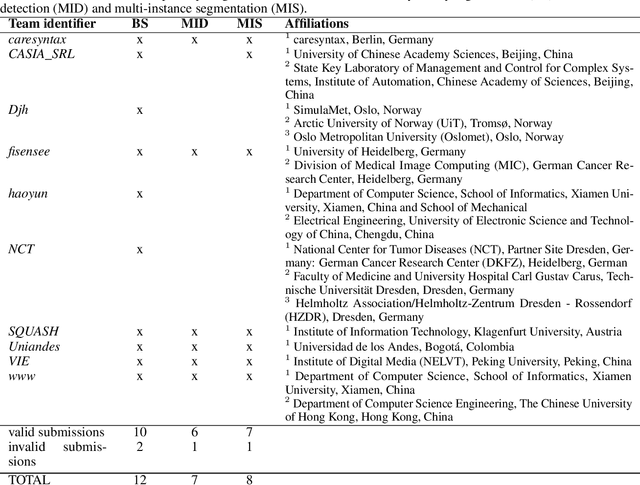
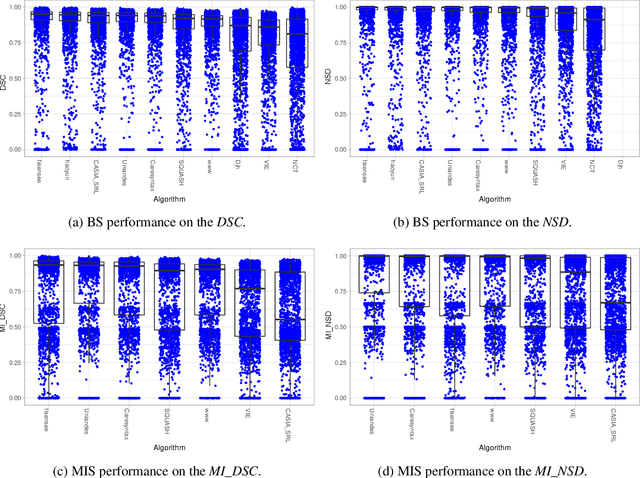
Abstract:Intraoperative tracking of laparoscopic instruments is often a prerequisite for computer and robotic-assisted interventions. While numerous methods for detecting, segmenting and tracking of medical instruments based on endoscopic video images have been proposed in the literature, key limitations remain to be addressed: Firstly, robustness, that is, the reliable performance of state-of-the-art methods when run on challenging images (e.g. in the presence of blood, smoke or motion artifacts). Secondly, generalization; algorithms trained for a specific intervention in a specific hospital should generalize to other interventions or institutions. In an effort to promote solutions for these limitations, we organized the Robust Medical Instrument Segmentation (ROBUST-MIS) challenge as an international benchmarking competition with a specific focus on the robustness and generalization capabilities of algorithms. For the first time in the field of endoscopic image processing, our challenge included a task on binary segmentation and also addressed multi-instance detection and segmentation. The challenge was based on a surgical data set comprising 10,040 annotated images acquired from a total of 30 surgical procedures from three different types of surgery. The validation of the competing methods for the three tasks (binary segmentation, multi-instance detection and multi-instance segmentation) was performed in three different stages with an increasing domain gap between the training and the test data. The results confirm the initial hypothesis, namely that algorithm performance degrades with an increasing domain gap. While the average detection and segmentation quality of the best-performing algorithms is high, future research should concentrate on detection and segmentation of small, crossing, moving and transparent instrument(s) (parts).
2018 Robotic Scene Segmentation Challenge
Feb 03, 2020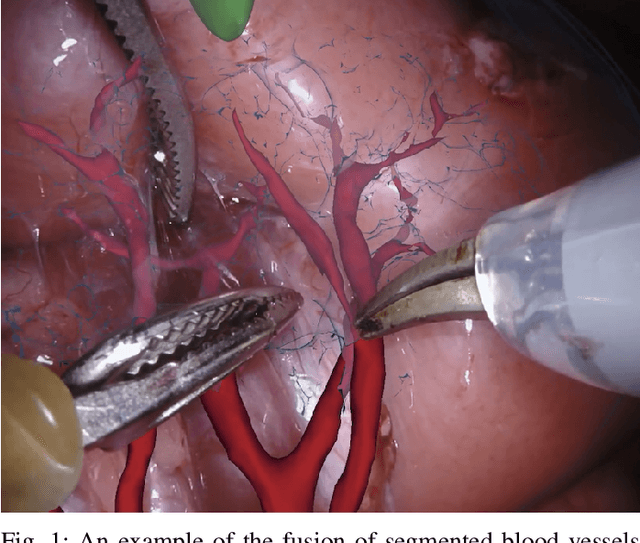
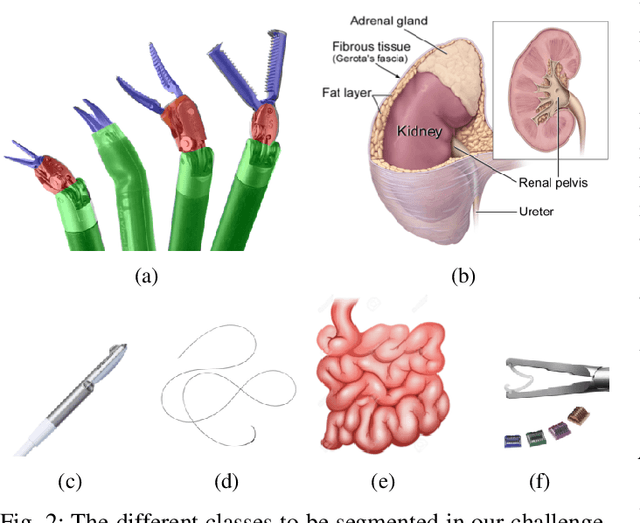
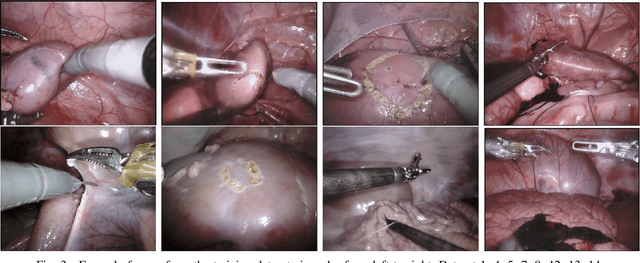
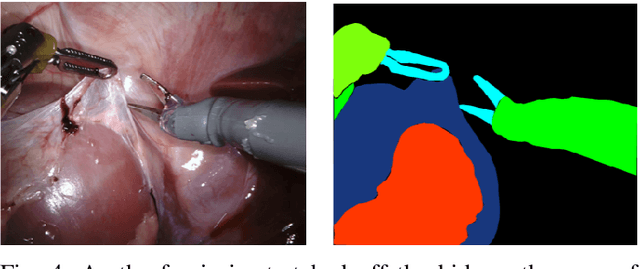
Abstract:In 2015 we began a sub-challenge at the EndoVis workshop at MICCAI in Munich using endoscope images of ex-vivo tissue with automatically generated annotations from robot forward kinematics and instrument CAD models. However, the limited background variation and simple motion rendered the dataset uninformative in learning about which techniques would be suitable for segmentation in real surgery. In 2017, at the same workshop in Quebec we introduced the robotic instrument segmentation dataset with 10 teams participating in the challenge to perform binary, articulating parts and type segmentation of da Vinci instruments. This challenge included realistic instrument motion and more complex porcine tissue as background and was widely addressed with modifications on U-Nets and other popular CNN architectures. In 2018 we added to the complexity by introducing a set of anatomical objects and medical devices to the segmented classes. To avoid over-complicating the challenge, we continued with porcine data which is dramatically simpler than human tissue due to the lack of fatty tissue occluding many organs.
Image biomarker standardisation initiative
Sep 17, 2018
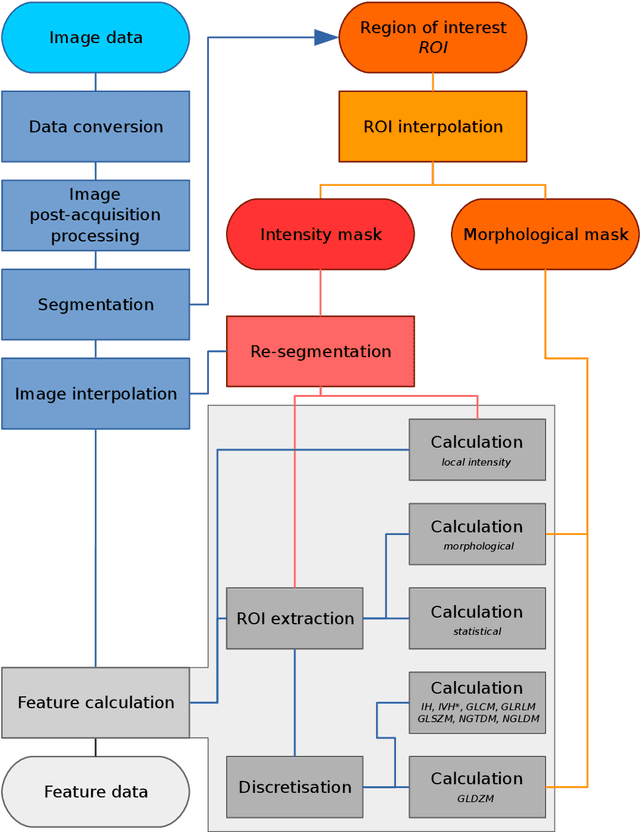
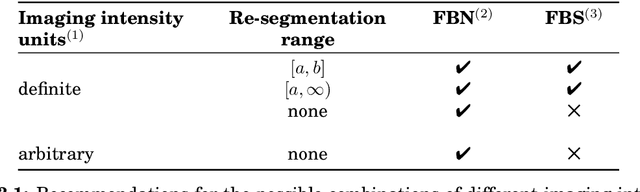
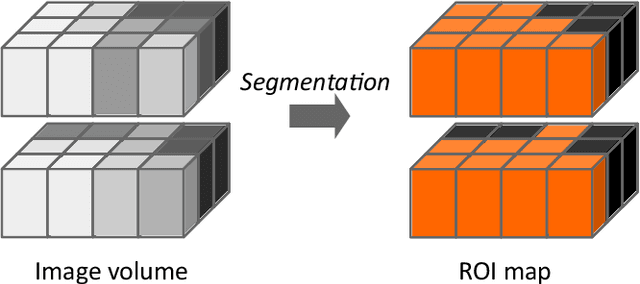
Abstract:The image biomarker standardisation initiative (IBSI) is an independent international collaboration which works towards standardising the extraction of image biomarkers from acquired imaging for the purpose of high-throughput quantitative image analysis (radiomics). Lack of reproducibility and validation of high-throughput quantitative image analysis studies is considered to be a major challenge for the field. Part of this challenge lies in the scantiness of consensus-based guidelines and definitions for the process of translating acquired imaging into high-throughput image biomarkers. The IBSI therefore seeks to provide image biomarker nomenclature and definitions, benchmark data sets, and benchmark values to verify image processing and image biomarker calculations, as well as reporting guidelines, for high-throughput image analysis.
Assessing robustness of radiomic features by image perturbation
Jun 18, 2018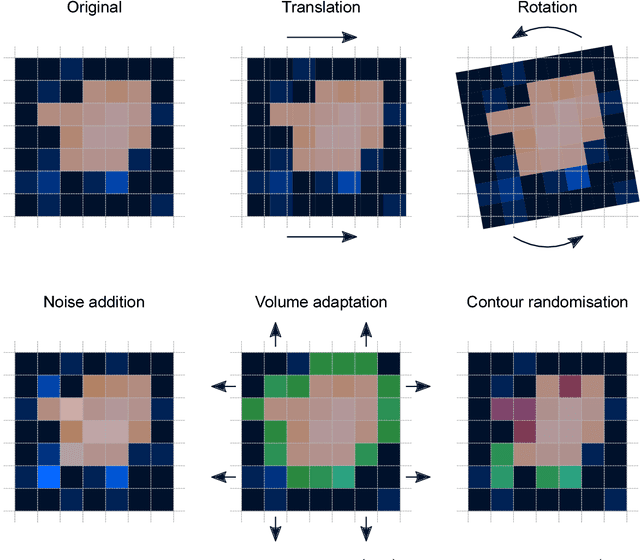
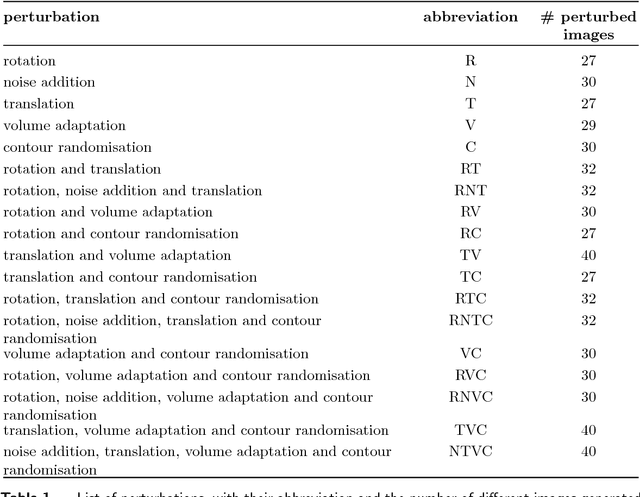
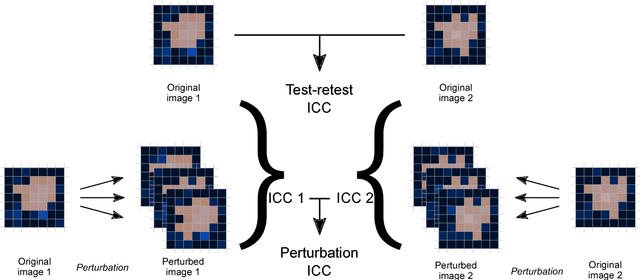
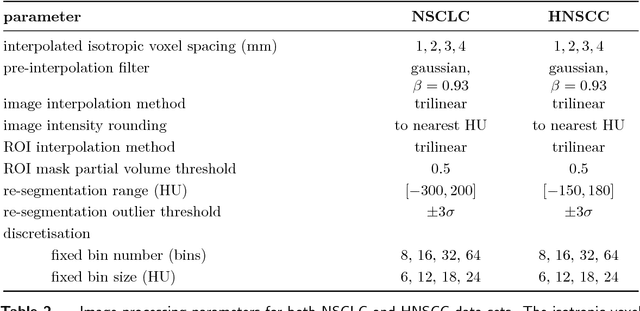
Abstract:Image features need to be robust against differences in positioning, acquisition and segmentation to ensure reproducibility. Radiomic models that only include robust features can be used to analyse new images, whereas models with non-robust features may fail to predict the outcome of interest accurately. Test-retest imaging is recommended to assess robustness, but may not be available for the phenotype of interest. We therefore investigated 18 methods to determine feature robustness based on image perturbations. Test-retest and perturbation robustness were compared for 4032 features that were computed from the gross tumour volume in two cohorts with computed tomography imaging: I) 31 non-small-cell lung cancer (NSCLC) patients; II): 19 head-and-neck squamous cell carcinoma (HNSCC) patients. Robustness was measured using the intraclass correlation coefficient (1,1) (ICC). Features with ICC$\geq0.90$ were considered robust. The NSCLC cohort contained more robust features for test-retest imaging than the HNSCC cohort ($73.5\%$ vs. $34.0\%$). A perturbation chain consisting of noise addition, affine translation, volume growth/shrinkage and supervoxel-based contour randomisation identified the fewest false positive robust features (NSCLC: $3.3\%$; HNSCC: $10.0\%$). Thus, this perturbation chain may be used to assess feature robustness.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge