Micha Pfeiffer
PIVOTS: Aligning unseen Structures using Preoperative to Intraoperative Volume-To-Surface Registration for Liver Navigation
Jul 27, 2025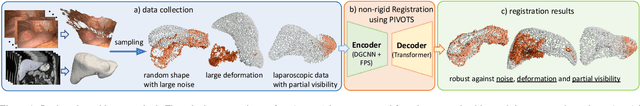
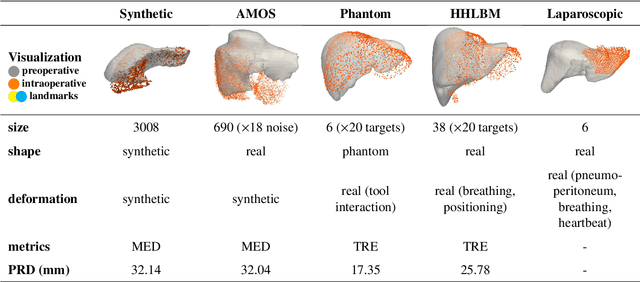
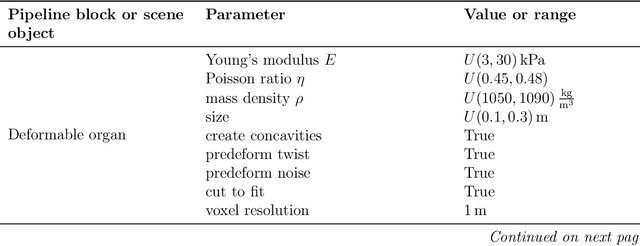
Abstract:Non-rigid registration is essential for Augmented Reality guided laparoscopic liver surgery by fusing preoperative information, such as tumor location and vascular structures, into the limited intraoperative view, thereby enhancing surgical navigation. A prerequisite is the accurate prediction of intraoperative liver deformation which remains highly challenging due to factors such as large deformation caused by pneumoperitoneum, respiration and tool interaction as well as noisy intraoperative data, and limited field of view due to occlusion and constrained camera movement. To address these challenges, we introduce PIVOTS, a Preoperative to Intraoperative VOlume-To-Surface registration neural network that directly takes point clouds as input for deformation prediction. The geometric feature extraction encoder allows multi-resolution feature extraction, and the decoder, comprising novel deformation aware cross attention modules, enables pre- and intraoperative information interaction and accurate multi-level displacement prediction. We train the neural network on synthetic data simulated from a biomechanical simulation pipeline and validate its performance on both synthetic and real datasets. Results demonstrate superior registration performance of our method compared to baseline methods, exhibiting strong robustness against high amounts of noise, large deformation, and various levels of intraoperative visibility. We publish the training and test sets as evaluation benchmarks and call for a fair comparison of liver registration methods with volume-to-surface data. Code and datasets are available here https://github.com/pengliu-nct/PIVOTS.
Mission Balance: Generating Under-represented Class Samples using Video Diffusion Models
May 14, 2025Abstract:Computer-assisted interventions can improve intra-operative guidance, particularly through deep learning methods that harness the spatiotemporal information in surgical videos. However, the severe data imbalance often found in surgical video datasets hinders the development of high-performing models. In this work, we aim to overcome the data imbalance by synthesizing surgical videos. We propose a unique two-stage, text-conditioned diffusion-based method to generate high-fidelity surgical videos for under-represented classes. Our approach conditions the generation process on text prompts and decouples spatial and temporal modeling by utilizing a 2D latent diffusion model to capture spatial content and then integrating temporal attention layers to ensure temporal consistency. Furthermore, we introduce a rejection sampling strategy to select the most suitable synthetic samples, effectively augmenting existing datasets to address class imbalance. We evaluate our method on two downstream tasks-surgical action recognition and intra-operative event prediction-demonstrating that incorporating synthetic videos from our approach substantially enhances model performance. We open-source our implementation at https://gitlab.com/nct_tso_public/surgvgen.
Synthesizing Multi-Class Surgical Datasets with Anatomy-Aware Diffusion Models
Oct 10, 2024Abstract:In computer-assisted surgery, automatically recognizing anatomical organs is crucial for understanding the surgical scene and providing intraoperative assistance. While machine learning models can identify such structures, their deployment is hindered by the need for labeled, diverse surgical datasets with anatomical annotations. Labeling multiple classes (i.e., organs) in a surgical scene is time-intensive, requiring medical experts. Although synthetically generated images can enhance segmentation performance, maintaining both organ structure and texture during generation is challenging. We introduce a multi-stage approach using diffusion models to generate multi-class surgical datasets with annotations. Our framework improves anatomy awareness by training organ specific models with an inpainting objective guided by binary segmentation masks. The organs are generated with an inference pipeline using pre-trained ControlNet to maintain the organ structure. The synthetic multi-class datasets are constructed through an image composition step, ensuring structural and textural consistency. This versatile approach allows the generation of multi-class datasets from real binary datasets and simulated surgical masks. We thoroughly evaluate the generated datasets on image quality and downstream segmentation, achieving a $15\%$ improvement in segmentation scores when combined with real images. Our codebase https://gitlab.com/nct_tso_public/muli-class-image-synthesis
SurgicaL-CD: Generating Surgical Images via Unpaired Image Translation with Latent Consistency Diffusion Models
Aug 19, 2024Abstract:Computer-assisted surgery (CAS) systems are designed to assist surgeons during procedures, thereby reducing complications and enhancing patient care. Training machine learning models for these systems requires a large corpus of annotated datasets, which is challenging to obtain in the surgical domain due to patient privacy concerns and the significant labeling effort required from doctors. Previous methods have explored unpaired image translation using generative models to create realistic surgical images from simulations. However, these approaches have struggled to produce high-quality, diverse surgical images. In this work, we introduce \emph{SurgicaL-CD}, a consistency-distilled diffusion method to generate realistic surgical images with only a few sampling steps without paired data. We evaluate our approach on three datasets, assessing the generated images in terms of quality and utility as downstream training datasets. Our results demonstrate that our method outperforms GANs and diffusion-based approaches. Our code is available at \url{https://gitlab.com/nct_tso_public/gan2diffusion}.
An objective comparison of methods for augmented reality in laparoscopic liver resection by preoperative-to-intraoperative image fusion
Feb 07, 2024



Abstract:Augmented reality for laparoscopic liver resection is a visualisation mode that allows a surgeon to localise tumours and vessels embedded within the liver by projecting them on top of a laparoscopic image. Preoperative 3D models extracted from CT or MRI data are registered to the intraoperative laparoscopic images during this process. In terms of 3D-2D fusion, most of the algorithms make use of anatomical landmarks to guide registration. These landmarks include the liver's inferior ridge, the falciform ligament, and the occluding contours. They are usually marked by hand in both the laparoscopic image and the 3D model, which is time-consuming and may contain errors if done by a non-experienced user. Therefore, there is a need to automate this process so that augmented reality can be used effectively in the operating room. We present the Preoperative-to-Intraoperative Laparoscopic Fusion Challenge (P2ILF), held during the Medical Imaging and Computer Assisted Interventions (MICCAI 2022) conference, which investigates the possibilities of detecting these landmarks automatically and using them in registration. The challenge was divided into two tasks: 1) A 2D and 3D landmark detection task and 2) a 3D-2D registration task. The teams were provided with training data consisting of 167 laparoscopic images and 9 preoperative 3D models from 9 patients, with the corresponding 2D and 3D landmark annotations. A total of 6 teams from 4 countries participated, whose proposed methods were evaluated on 16 images and two preoperative 3D models from two patients. All the teams proposed deep learning-based methods for the 2D and 3D landmark segmentation tasks and differentiable rendering-based methods for the registration task. Based on the experimental outcomes, we propose three key hypotheses that determine current limitations and future directions for research in this domain.
Exploring Semantic Consistency in Unpaired Image Translation to Generate Data for Surgical Applications
Sep 11, 2023Abstract:In surgical computer vision applications, obtaining labeled training data is challenging due to data-privacy concerns and the need for expert annotation. Unpaired image-to-image translation techniques have been explored to automatically generate large annotated datasets by translating synthetic images to the realistic domain. However, preserving the structure and semantic consistency between the input and translated images presents significant challenges, mainly when there is a distributional mismatch in the semantic characteristics of the domains. This study empirically investigates unpaired image translation methods for generating suitable data in surgical applications, explicitly focusing on semantic consistency. We extensively evaluate various state-of-the-art image translation models on two challenging surgical datasets and downstream semantic segmentation tasks. We find that a simple combination of structural-similarity loss and contrastive learning yields the most promising results. Quantitatively, we show that the data generated with this approach yields higher semantic consistency and can be used more effectively as training data.
Long-Term Temporally Consistent Unpaired Video Translation from Simulated Surgical 3D Data
Mar 31, 2021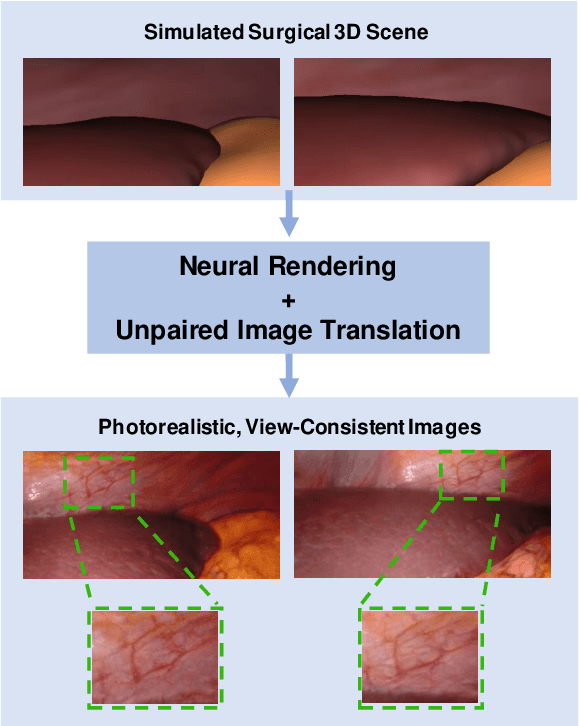
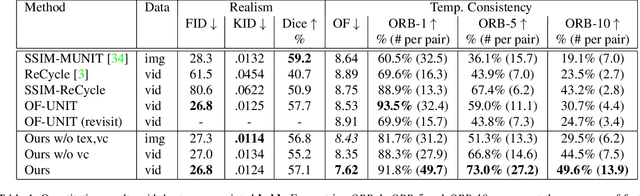
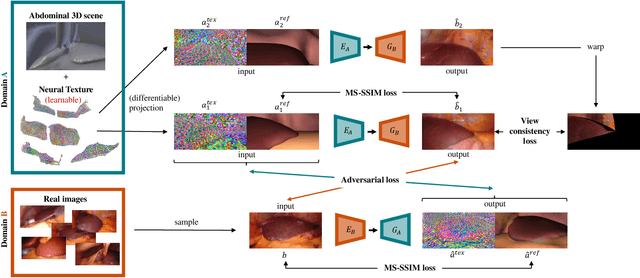
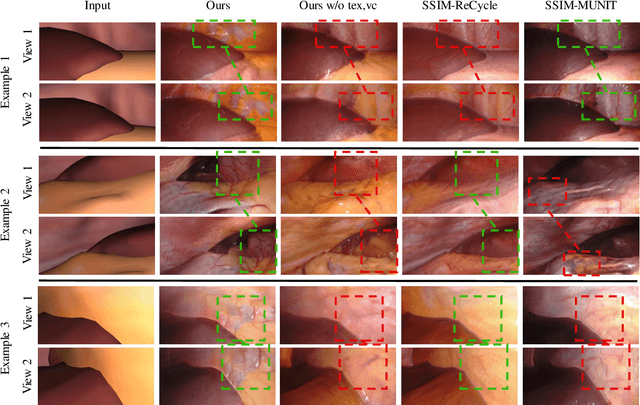
Abstract:Research in unpaired video translation has mainly focused on short-term temporal consistency by conditioning on neighboring frames. However for transfer from simulated to photorealistic sequences, available information on the underlying geometry offers potential for achieving global consistency across views. We propose a novel approach which combines unpaired image translation with neural rendering to transfer simulated to photorealistic surgical abdominal scenes. By introducing global learnable textures and a lighting-invariant view-consistency loss, our method produces consistent translations of arbitrary views and thus enables long-term consistent video synthesis. We design and test our model to generate video sequences from minimally-invasive surgical abdominal scenes. Because labeled data is often limited in this domain, photorealistic data where ground truth information from the simulated domain is preserved is especially relevant. By extending existing image-based methods to view-consistent videos, we aim to impact the applicability of simulated training and evaluation environments for surgical applications. Code and data will be made publicly available soon.
Non-Rigid Volume to Surface Registration using a Data-Driven Biomechanical Model
May 29, 2020



Abstract:Non-rigid registration is a key component in soft-tissue navigation. We focus on laparoscopic liver surgery, where we register the organ model obtained from a preoperative CT scan to the intraoperative partial organ surface, reconstructed from the laparoscopic video. This is a challenging task due to sparse and noisy intraoperative data, real-time requirements and many unknowns - such as tissue properties and boundary conditions. Furthermore, establishing correspondences between pre- and intraoperative data can be extremely difficult since the liver usually lacks distinct surface features and the used imaging modalities suffer from very different types of noise. In this work, we train a convolutional neural network to perform both the search for surface correspondences as well as the non-rigid registration in one step. The network is trained on physically accurate biomechanical simulations of randomly generated, deforming organ-like structures. This enables the network to immediately generalize to a new patient organ without the need to re-train. We add various amounts of noise to the intraoperative surfaces during training, making the network robust to noisy intraoperative data. During inference, the network outputs the displacement field which matches the preoperative volume to the partial intraoperative surface. In multiple experiments, we show that the network translates well to real data while maintaining a high inference speed. Our code is made available online.
Generating large labeled data sets for laparoscopic image processing tasks using unpaired image-to-image translation
Jul 05, 2019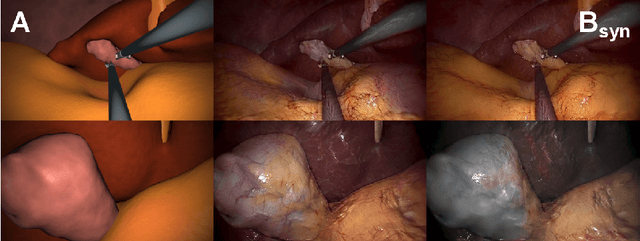
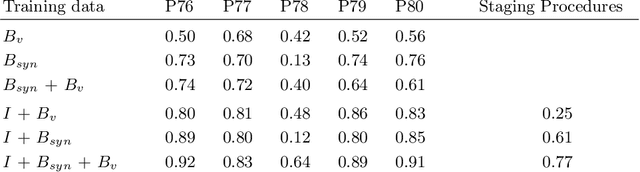
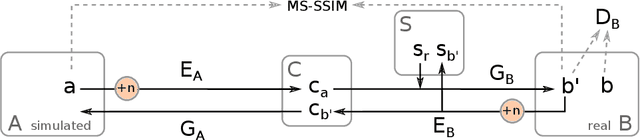
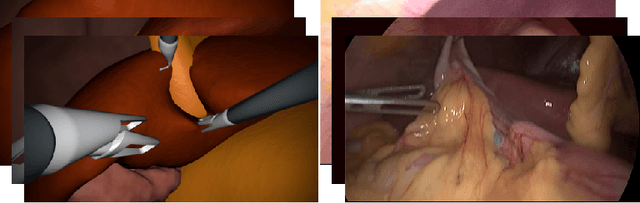
Abstract:In the medical domain, the lack of large training data sets and benchmarks is often a limiting factor for training deep neural networks. In contrast to expensive manual labeling, computer simulations can generate large and fully labeled data sets with a minimum of manual effort. However, models that are trained on simulated data usually do not translate well to real scenarios. To bridge the domain gap between simulated and real laparoscopic images, we exploit recent advances in unpaired image-to-image translation. We extent an image-to-image translation method to generate a diverse multitude of realistically looking synthetic images based on images from a simple laparoscopy simulation. By incorporating means to ensure that the image content is preserved during the translation process, we ensure that the labels given for the simulated images remain valid for their realistically looking translations. This way, we are able to generate a large, fully labeled synthetic data set of laparoscopic images with realistic appearance. We show that this data set can be used to train models for the task of liver segmentation of laparoscopic images. We achieve average dice scores of up to 0.89 in some patients without manually labeling a single laparoscopic image and show that using our synthetic data to pre-train models can greatly improve their performance. The synthetic data set will be made publicly available, fully labeled with segmentation maps, depth maps, normal maps, and positions of tools and camera (http://opencas.dkfz.de/image2image).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge