Stefanie Speidel
*: shared first/last authors
A benchmark for video-based laparoscopic skill analysis and assessment
Feb 10, 2026Abstract:Laparoscopic surgery is a complex surgical technique that requires extensive training. Recent advances in deep learning have shown promise in supporting this training by enabling automatic video-based assessment of surgical skills. However, the development and evaluation of deep learning models is currently hindered by the limited size of available annotated datasets. To address this gap, we introduce the Laparoscopic Skill Analysis and Assessment (LASANA) dataset, comprising 1270 stereo video recordings of four basic laparoscopic training tasks. Each recording is annotated with a structured skill rating, aggregated from three independent raters, as well as binary labels indicating the presence or absence of task-specific errors. The majority of recordings originate from a laparoscopic training course, thereby reflecting a natural variation in the skill of participants. To facilitate benchmarking of both existing and novel approaches for video-based skill assessment and error recognition, we provide predefined data splits for each task. Furthermore, we present baseline results from a deep learning model as a reference point for future comparisons.
Adaptive-CaRe: Adaptive Causal Regularization for Robust Outcome Prediction
Feb 06, 2026Abstract:Accurate prediction of outcomes is crucial for clinical decision-making and personalized patient care. Supervised machine learning algorithms, which are commonly used for outcome prediction in the medical domain, optimize for predictive accuracy, which can result in models latching onto spurious correlations instead of robust predictors. Causal structure learning methods on the other hand have the potential to provide robust predictors for the target, but can be too conservative because of algorithmic and data assumptions, resulting in loss of diagnostic precision. Therefore, we propose a novel model-agnostic regularization strategy, Adaptive-CaRe, for generalized outcome prediction in the medical domain. Adaptive-CaRe strikes a balance between both predictive value and causal robustness by incorporating a penalty that is proportional to the difference between the estimated statistical contribution and estimated causal contribution of the input features for model predictions. Our experiments on synthetic data establish the efficacy of the proposed Adaptive-CaRe regularizer in finding robust predictors for the target while maintaining competitive predictive accuracy. With experiments on a standard causal benchmark, we provide a blueprint for navigating the trade-off between predictive accuracy and causal robustness by tweaking the regularization strength, $λ$. Validation using real-world dataset confirms that the results translate to practical, real-domain settings. Therefore, Adaptive-CaRe provides a simple yet effective solution to the long-standing trade-off between predictive accuracy and causal robustness in the medical domain. Future work would involve studying alternate causal structure learning frameworks and complex classification models to provide deeper insights at a larger scale.
MoLF: Mixture-of-Latent-Flow for Pan-Cancer Spatial Gene Expression Prediction from Histology
Feb 02, 2026Abstract:Inferring spatial transcriptomics (ST) from histology enables scalable histogenomic profiling, yet current methods are largely restricted to single-tissue models. This fragmentation fails to leverage biological principles shared across cancer types and hinders application to data-scarce scenarios. While pan-cancer training offers a solution, the resulting heterogeneity challenges monolithic architectures. To bridge this gap, we introduce MoLF (Mixture-of-Latent-Flow), a generative model for pan-cancer histogenomic prediction. MoLF leverages a conditional Flow Matching objective to map noise to the gene latent manifold, parameterized by a Mixture-of-Experts (MoE) velocity field. By dynamically routing inputs to specialized sub-networks, this architecture effectively decouples the optimization of diverse tissue patterns. Our experiments demonstrate that MoLF establishes a new state-of-the-art, consistently outperforming both specialized and foundation model baselines on pan-cancer benchmarks. Furthermore, MoLF exhibits zero-shot generalization to cross-species data, suggesting it captures fundamental, conserved histo-molecular mechanisms.
MoE-ACT: Improving Surgical Imitation Learning Policies through Supervised Mixture-of-Experts
Jan 29, 2026Abstract:Imitation learning has achieved remarkable success in robotic manipulation, yet its application to surgical robotics remains challenging due to data scarcity, constrained workspaces, and the need for an exceptional level of safety and predictability. We present a supervised Mixture-of-Experts (MoE) architecture designed for phase-structured surgical manipulation tasks, which can be added on top of any autonomous policy. Unlike prior surgical robot learning approaches that rely on multi-camera setups or thousands of demonstrations, we show that a lightweight action decoder policy like Action Chunking Transformer (ACT) can learn complex, long-horizon manipulation from less than 150 demonstrations using solely stereo endoscopic images, when equipped with our architecture. We evaluate our approach on the collaborative surgical task of bowel grasping and retraction, where a robot assistant interprets visual cues from a human surgeon, executes targeted grasping on deformable tissue, and performs sustained retraction. We benchmark our method against state-of-the-art Vision-Language-Action (VLA) models and the standard ACT baseline. Our results show that generalist VLAs fail to acquire the task entirely, even under standard in-distribution conditions. Furthermore, while standard ACT achieves moderate success in-distribution, adopting a supervised MoE architecture significantly boosts its performance, yielding higher success rates in-distribution and demonstrating superior robustness in out-of-distribution scenarios, including novel grasp locations, reduced illumination, and partial occlusions. Notably, it generalizes to unseen testing viewpoints and also transfers zero-shot to ex vivo porcine tissue without additional training, offering a promising pathway toward in vivo deployment. To support this, we present qualitative preliminary results of policy roll-outs during in vivo porcine surgery.
HistoPrism: Unlocking Functional Pathway Analysis from Pan-Cancer Histology via Gene Expression Prediction
Jan 29, 2026Abstract:Predicting spatial gene expression from H&E histology offers a scalable and clinically accessible alternative to sequencing, but realizing clinical impact requires models that generalize across cancer types and capture biologically coherent signals. Prior work is often limited to per-cancer settings and variance-based evaluation, leaving functional relevance underexplored. We introduce HistoPrism, an efficient transformer-based architecture for pan-cancer prediction of gene expression from histology. To evaluate biological meaning, we introduce a pathway-level benchmark, shifting assessment from isolated gene-level variance to coherent functional pathways. HistoPrism not only surpasses prior state-of-the-art models on highly variable genes , but also more importantly, achieves substantial gains on pathway-level prediction, demonstrating its ability to recover biologically coherent transcriptomic patterns. With strong pan-cancer generalization and improved efficiency, HistoPrism establishes a new standard for clinically relevant transcriptomic modeling from routinely available histology.
* Accepted at ICLR2026
Federated Learning for Surgical Vision in Appendicitis Classification: Results of the FedSurg EndoVis 2024 Challenge
Oct 06, 2025Abstract:Purpose: The FedSurg challenge was designed to benchmark the state of the art in federated learning for surgical video classification. Its goal was to assess how well current methods generalize to unseen clinical centers and adapt through local fine-tuning while enabling collaborative model development without sharing patient data. Methods: Participants developed strategies to classify inflammation stages in appendicitis using a preliminary version of the multi-center Appendix300 video dataset. The challenge evaluated two tasks: generalization to an unseen center and center-specific adaptation after fine-tuning. Submitted approaches included foundation models with linear probing, metric learning with triplet loss, and various FL aggregation schemes (FedAvg, FedMedian, FedSAM). Performance was assessed using F1-score and Expected Cost, with ranking robustness evaluated via bootstrapping and statistical testing. Results: In the generalization task, performance across centers was limited. In the adaptation task, all teams improved after fine-tuning, though ranking stability was low. The ViViT-based submission achieved the strongest overall performance. The challenge highlighted limitations in generalization, sensitivity to class imbalance, and difficulties in hyperparameter tuning in decentralized training, while spatiotemporal modeling and context-aware preprocessing emerged as promising strategies. Conclusion: The FedSurg Challenge establishes the first benchmark for evaluating FL strategies in surgical video classification. Findings highlight the trade-off between local personalization and global robustness, and underscore the importance of architecture choice, preprocessing, and loss design. This benchmarking offers a reference point for future development of imbalance-aware, adaptive, and robust FL methods in clinical surgical AI.
Vibration-Based Energy Metric for Restoring Needle Alignment in Autonomous Robotic Ultrasound
Aug 09, 2025Abstract:Precise needle alignment is essential for percutaneous needle insertion in robotic ultrasound-guided procedures. However, inherent challenges such as speckle noise, needle-like artifacts, and low image resolution make robust needle detection difficult, particularly when visibility is reduced or lost. In this paper, we propose a method to restore needle alignment when the ultrasound imaging plane and the needle insertion plane are misaligned. Unlike many existing approaches that rely heavily on needle visibility in ultrasound images, our method uses a more robust feature by periodically vibrating the needle using a mechanical system. Specifically, we propose a vibration-based energy metric that remains effective even when the needle is fully out of plane. Using this metric, we develop a control strategy to reposition the ultrasound probe in response to misalignments between the imaging plane and the needle insertion plane in both translation and rotation. Experiments conducted on ex-vivo porcine tissue samples using a dual-arm robotic ultrasound-guided needle insertion system demonstrate the effectiveness of the proposed approach. The experimental results show the translational error of 0.41$\pm$0.27 mm and the rotational error of 0.51$\pm$0.19 degrees.
PIVOTS: Aligning unseen Structures using Preoperative to Intraoperative Volume-To-Surface Registration for Liver Navigation
Jul 27, 2025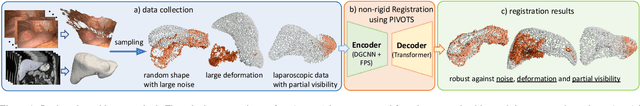
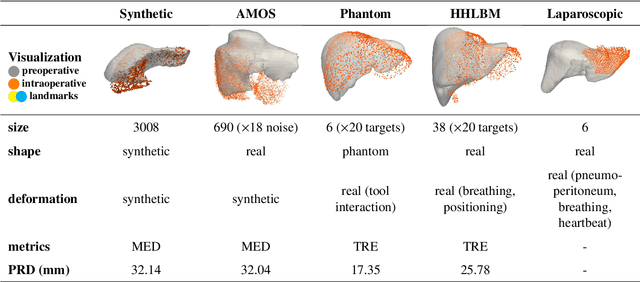
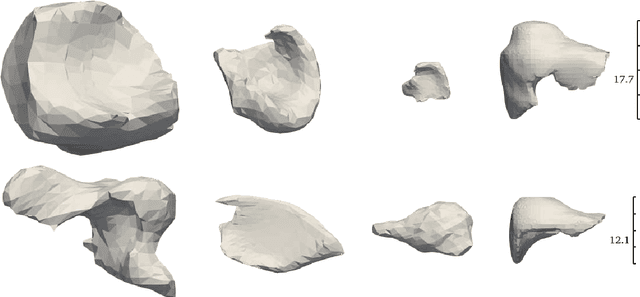
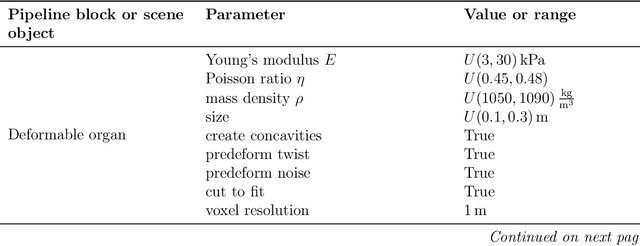
Abstract:Non-rigid registration is essential for Augmented Reality guided laparoscopic liver surgery by fusing preoperative information, such as tumor location and vascular structures, into the limited intraoperative view, thereby enhancing surgical navigation. A prerequisite is the accurate prediction of intraoperative liver deformation which remains highly challenging due to factors such as large deformation caused by pneumoperitoneum, respiration and tool interaction as well as noisy intraoperative data, and limited field of view due to occlusion and constrained camera movement. To address these challenges, we introduce PIVOTS, a Preoperative to Intraoperative VOlume-To-Surface registration neural network that directly takes point clouds as input for deformation prediction. The geometric feature extraction encoder allows multi-resolution feature extraction, and the decoder, comprising novel deformation aware cross attention modules, enables pre- and intraoperative information interaction and accurate multi-level displacement prediction. We train the neural network on synthetic data simulated from a biomechanical simulation pipeline and validate its performance on both synthetic and real datasets. Results demonstrate superior registration performance of our method compared to baseline methods, exhibiting strong robustness against high amounts of noise, large deformation, and various levels of intraoperative visibility. We publish the training and test sets as evaluation benchmarks and call for a fair comparison of liver registration methods with volume-to-surface data. Code and datasets are available here https://github.com/pengliu-nct/PIVOTS.
Comparative validation of surgical phase recognition, instrument keypoint estimation, and instrument instance segmentation in endoscopy: Results of the PhaKIR 2024 challenge
Jul 22, 2025Abstract:Reliable recognition and localization of surgical instruments in endoscopic video recordings are foundational for a wide range of applications in computer- and robot-assisted minimally invasive surgery (RAMIS), including surgical training, skill assessment, and autonomous assistance. However, robust performance under real-world conditions remains a significant challenge. Incorporating surgical context - such as the current procedural phase - has emerged as a promising strategy to improve robustness and interpretability. To address these challenges, we organized the Surgical Procedure Phase, Keypoint, and Instrument Recognition (PhaKIR) sub-challenge as part of the Endoscopic Vision (EndoVis) challenge at MICCAI 2024. We introduced a novel, multi-center dataset comprising thirteen full-length laparoscopic cholecystectomy videos collected from three distinct medical institutions, with unified annotations for three interrelated tasks: surgical phase recognition, instrument keypoint estimation, and instrument instance segmentation. Unlike existing datasets, ours enables joint investigation of instrument localization and procedural context within the same data while supporting the integration of temporal information across entire procedures. We report results and findings in accordance with the BIAS guidelines for biomedical image analysis challenges. The PhaKIR sub-challenge advances the field by providing a unique benchmark for developing temporally aware, context-driven methods in RAMIS and offers a high-quality resource to support future research in surgical scene understanding.
Mission Balance: Generating Under-represented Class Samples using Video Diffusion Models
May 14, 2025Abstract:Computer-assisted interventions can improve intra-operative guidance, particularly through deep learning methods that harness the spatiotemporal information in surgical videos. However, the severe data imbalance often found in surgical video datasets hinders the development of high-performing models. In this work, we aim to overcome the data imbalance by synthesizing surgical videos. We propose a unique two-stage, text-conditioned diffusion-based method to generate high-fidelity surgical videos for under-represented classes. Our approach conditions the generation process on text prompts and decouples spatial and temporal modeling by utilizing a 2D latent diffusion model to capture spatial content and then integrating temporal attention layers to ensure temporal consistency. Furthermore, we introduce a rejection sampling strategy to select the most suitable synthetic samples, effectively augmenting existing datasets to address class imbalance. We evaluate our method on two downstream tasks-surgical action recognition and intra-operative event prediction-demonstrating that incorporating synthetic videos from our approach substantially enhances model performance. We open-source our implementation at https://gitlab.com/nct_tso_public/surgvgen.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge