Fiona R. Kolbinger
Field strength-dependent performance variability in deep learning-based analysis of magnetic resonance imaging
Dec 18, 2025Abstract:This study quantitatively evaluates the impact of MRI scanner magnetic field strength on the performance and generalizability of deep learning-based segmentation algorithms. Three publicly available MRI datasets (breast tumor, pancreas, and cervical spine) were stratified by scanner field strength (1.5T vs. 3.0T). For each segmentation task, three nnU-Net-based models were developed: A model trained on 1.5T data only (m-1.5T), a model trained on 3.0T data only (m-3.0T), and a model trained on pooled 1.5T and 3.0T data (m-combined). Each model was evaluated on both 1.5T and 3.0T validation sets. Field-strength-dependent performance differences were investigated via Uniform Manifold Approximation and Projection (UMAP)-based clustering and radiomic analysis, including 23 first-order and texture features. For breast tumor segmentation, m-3.0T (DSC: 0.494 [1.5T] and 0.433 [3.0T]) significantly outperformed m-1.5T (DSC: 0.411 [1.5T] and 0.289 [3.0T]) and m-combined (DSC: 0.373 [1.5T] and 0.268[3.0T]) on both validation sets (p<0.0001). Pancreas segmentation showed similar trends: m-3.0T achieved the highest DSC (0.774 [1.5T], 0.840 [3.0T]), while m-1.5T underperformed significantly (p<0.0001). For cervical spine, models performed optimally on same-field validation sets with minimal cross-field performance degradation (DSC>0.92 for all comparisons). Radiomic analysis revealed moderate field-strength-dependent clustering in soft tissues (silhouette scores 0.23-0.29) but minimal separation in osseous structures (0.12). These results indicate that magnetic field strength in the training data substantially influences the performance of deep learning-based segmentation models, particularly for soft-tissue structures (e.g., small lesions). This warrants consideration of magnetic field strength as a confounding factor in studies evaluating AI performance on MRI.
Federated Learning for Surgical Vision in Appendicitis Classification: Results of the FedSurg EndoVis 2024 Challenge
Oct 06, 2025Abstract:Purpose: The FedSurg challenge was designed to benchmark the state of the art in federated learning for surgical video classification. Its goal was to assess how well current methods generalize to unseen clinical centers and adapt through local fine-tuning while enabling collaborative model development without sharing patient data. Methods: Participants developed strategies to classify inflammation stages in appendicitis using a preliminary version of the multi-center Appendix300 video dataset. The challenge evaluated two tasks: generalization to an unseen center and center-specific adaptation after fine-tuning. Submitted approaches included foundation models with linear probing, metric learning with triplet loss, and various FL aggregation schemes (FedAvg, FedMedian, FedSAM). Performance was assessed using F1-score and Expected Cost, with ranking robustness evaluated via bootstrapping and statistical testing. Results: In the generalization task, performance across centers was limited. In the adaptation task, all teams improved after fine-tuning, though ranking stability was low. The ViViT-based submission achieved the strongest overall performance. The challenge highlighted limitations in generalization, sensitivity to class imbalance, and difficulties in hyperparameter tuning in decentralized training, while spatiotemporal modeling and context-aware preprocessing emerged as promising strategies. Conclusion: The FedSurg Challenge establishes the first benchmark for evaluating FL strategies in surgical video classification. Findings highlight the trade-off between local personalization and global robustness, and underscore the importance of architecture choice, preprocessing, and loss design. This benchmarking offers a reference point for future development of imbalance-aware, adaptive, and robust FL methods in clinical surgical AI.
Federated EndoViT: Pretraining Vision Transformers via Federated Learning on Endoscopic Image Collections
Apr 23, 2025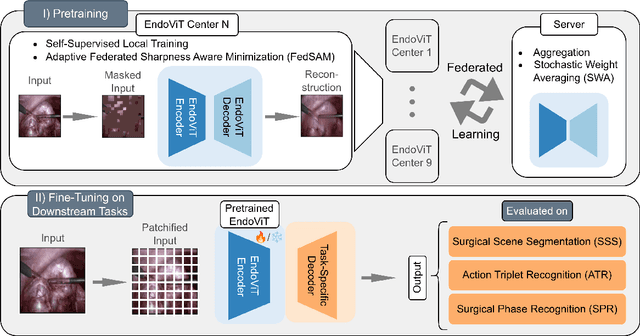
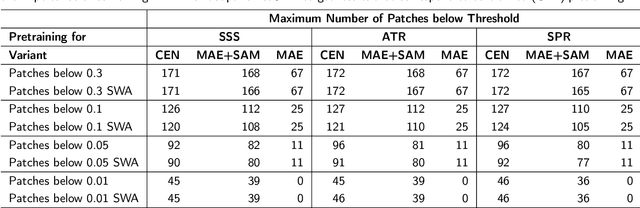
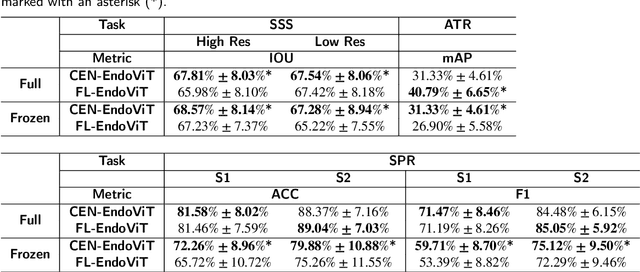
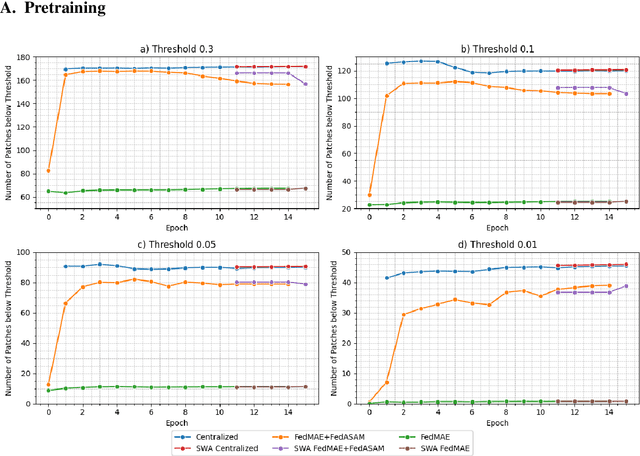
Abstract:Purpose: In this study, we investigate the training of foundation models using federated learning to address data-sharing limitations and enable collaborative model training without data transfer for minimally invasive surgery. Methods: Inspired by the EndoViT study, we adapt the Masked Autoencoder for federated learning, enhancing it with adaptive Sharpness-Aware Minimization (FedSAM) and Stochastic Weight Averaging (SWA). Our model is pretrained on the Endo700k dataset collection and later fine-tuned and evaluated for tasks such as Semantic Segmentation, Action Triplet Recognition, and Surgical Phase Recognition. Results: Our findings demonstrate that integrating adaptive FedSAM into the federated MAE approach improves pretraining, leading to a reduction in reconstruction loss per patch. The application of FL-EndoViT in surgical downstream tasks results in performance comparable to CEN-EndoViT. Furthermore, FL-EndoViT exhibits advantages over CEN-EndoViT in surgical scene segmentation when data is limited and in action triplet recognition when large datasets are used. Conclusion: These findings highlight the potential of federated learning for privacy-preserving training of surgical foundation models, offering a robust and generalizable solution for surgical data science. Effective collaboration requires adapting federated learning methods, such as the integration of FedSAM, which can accommodate the inherent data heterogeneity across institutions. In future, exploring FL in video-based models may enhance these capabilities by incorporating spatiotemporal dynamics crucial for real-world surgical environments.
Strategies to Improve Real-World Applicability of Laparoscopic Anatomy Segmentation Models
Mar 25, 2024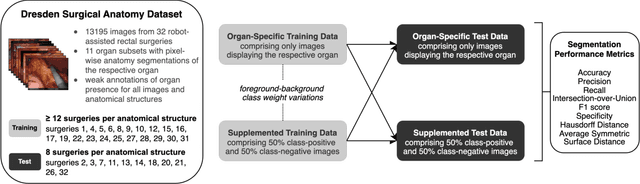


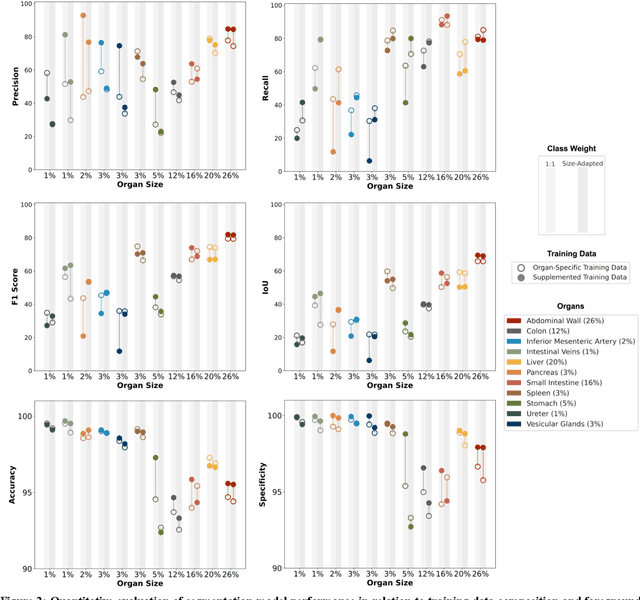
Abstract:Accurate identification and localization of anatomical structures of varying size and appearance in laparoscopic imaging are necessary to leverage the potential of computer vision techniques for surgical decision support. Segmentation performance of such models is traditionally reported using metrics of overlap such as IoU. However, imbalanced and unrealistic representation of classes in the training data and suboptimal selection of reported metrics have the potential to skew nominal segmentation performance and thereby ultimately limit clinical translation. In this work, we systematically analyze the impact of class characteristics (i.e., organ size differences), training and test data composition (i.e., representation of positive and negative examples), and modeling parameters (i.e., foreground-to-background class weight) on eight segmentation metrics: accuracy, precision, recall, IoU, F1 score, specificity, Hausdorff Distance, and Average Symmetric Surface Distance. Based on our findings, we propose two simple yet effective strategies to improve real-world applicability of image segmentation models in laparoscopic surgical data: (1) inclusion of negative examples in the training process and (2) adaptation of foreground-background weights in segmentation models to maximize model performance with respect to specific metrics of interest, depending on the clinical use case.
One model to use them all: Training a segmentation model with complementary datasets
Feb 29, 2024Abstract:Understanding a surgical scene is crucial for computer-assisted surgery systems to provide any intelligent assistance functionality. One way of achieving this scene understanding is via scene segmentation, where every pixel of a frame is classified and therefore identifies the visible structures and tissues. Progress on fully segmenting surgical scenes has been made using machine learning. However, such models require large amounts of annotated training data, containing examples of all relevant object classes. Such fully annotated datasets are hard to create, as every pixel in a frame needs to be annotated by medical experts and, therefore, are rarely available. In this work, we propose a method to combine multiple partially annotated datasets, which provide complementary annotations, into one model, enabling better scene segmentation and the use of multiple readily available datasets. Our method aims to combine available data with complementary labels by leveraging mutual exclusive properties to maximize information. Specifically, we propose to use positive annotations of other classes as negative samples and to exclude background pixels of binary annotations, as we cannot tell if they contain a class not annotated but predicted by the model. We evaluate our method by training a DeepLabV3 on the publicly available Dresden Surgical Anatomy Dataset, which provides multiple subsets of binary segmented anatomical structures. Our approach successfully combines 6 classes into one model, increasing the overall Dice Score by 4.4% compared to an ensemble of models trained on the classes individually. By including information on multiple classes, we were able to reduce confusion between stomach and colon by 24%. Our results demonstrate the feasibility of training a model on multiple datasets. This paves the way for future work further alleviating the need for one large, fully segmented datasets.
Regression-based Deep-Learning predicts molecular biomarkers from pathology slides
Apr 11, 2023Abstract:Deep Learning (DL) can predict biomarkers from cancer histopathology. Several clinically approved applications use this technology. Most approaches, however, predict categorical labels, whereas biomarkers are often continuous measurements. We hypothesized that regression-based DL outperforms classification-based DL. Therefore, we developed and evaluated a new self-supervised attention-based weakly supervised regression method that predicts continuous biomarkers directly from images in 11,671 patients across nine cancer types. We tested our method for multiple clinically and biologically relevant biomarkers: homologous repair deficiency (HRD) score, a clinically used pan-cancer biomarker, as well as markers of key biological processes in the tumor microenvironment. Using regression significantly enhances the accuracy of biomarker prediction, while also improving the interpretability of the results over classification. In a large cohort of colorectal cancer patients, regression-based prediction scores provide a higher prognostic value than classification-based scores. Our open-source regression approach offers a promising alternative for continuous biomarker analysis in computational pathology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge