Omar S. M. El Nahhas
Abnormality-Driven Representation Learning for Radiology Imaging
Nov 25, 2024



Abstract:To date, the most common approach for radiology deep learning pipelines is the use of end-to-end 3D networks based on models pre-trained on other tasks, followed by fine-tuning on the task at hand. In contrast, adjacent medical fields such as pathology, which focus on 2D images, have effectively adopted task-agnostic foundational models based on self-supervised learning (SSL), combined with weakly-supervised deep learning (DL). However, the field of radiology still lacks task-agnostic representation models due to the computational and data demands of 3D imaging and the anatomical complexity inherent to radiology scans. To address this gap, we propose CLEAR, a framework for radiology images that uses extracted embeddings from 2D slices along with attention-based aggregation for efficiently predicting clinical endpoints. As part of this framework, we introduce lesion-enhanced contrastive learning (LeCL), a novel approach to obtain visual representations driven by abnormalities in 2D axial slices across different locations of the CT scans. Specifically, we trained single-domain contrastive learning approaches using three different architectures: Vision Transformers, Vision State Space Models and Gated Convolutional Neural Networks. We evaluate our approach across three clinical tasks: tumor lesion location, lung disease detection, and patient staging, benchmarking against four state-of-the-art foundation models, including BiomedCLIP. Our findings demonstrate that CLEAR using representations learned through LeCL, outperforms existing foundation models, while being substantially more compute- and data-efficient.
Benchmarking foundation models as feature extractors for weakly-supervised computational pathology
Aug 28, 2024Abstract:Advancements in artificial intelligence have driven the development of numerous pathology foundation models capable of extracting clinically relevant information. However, there is currently limited literature independently evaluating these foundation models on truly external cohorts and clinically-relevant tasks to uncover adjustments for future improvements. In this study, we benchmarked ten histopathology foundation models on 13 patient cohorts with 6,791 patients and 9,493 slides from lung, colorectal, gastric, and breast cancers. The models were evaluated on weakly-supervised tasks related to biomarkers, morphological properties, and prognostic outcomes. We show that a vision-language foundation model, CONCH, yielded the highest performance in 42% of tasks when compared to vision-only foundation models. The experiments reveal that foundation models trained on distinct cohorts learn complementary features to predict the same label, and can be fused to outperform the current state of the art. Creating an ensemble of complementary foundation models outperformed CONCH in 66% of tasks. Moreover, our findings suggest that data diversity outweighs data volume for foundation models. Our work highlights actionable adjustments to improve pathology foundation models.
Compute-Efficient Medical Image Classification with Softmax-Free Transformers and Sequence Normalization
Jun 03, 2024Abstract:The Transformer model has been pivotal in advancing fields such as natural language processing, speech recognition, and computer vision. However, a critical limitation of this model is its quadratic computational and memory complexity relative to the sequence length, which constrains its application to longer sequences. This is especially crucial in medical imaging where high-resolution images can reach gigapixel scale. Efforts to address this issue have predominantely focused on complex techniques, such as decomposing the softmax operation integral to the Transformer's architecture. This paper addresses this quadratic computational complexity of Transformer models and introduces a remarkably simple and effective method that circumvents this issue by eliminating the softmax function from the attention mechanism and adopting a sequence normalization technique for the key, query, and value tokens. Coupled with a reordering of matrix multiplications this approach reduces the memory- and compute complexity to a linear scale. We evaluate this approach across various medical imaging datasets comprising fundoscopic, dermascopic, radiologic and histologic imaging data. Our findings highlight that these models exhibit a comparable performance to traditional transformer models, while efficiently handling longer sequences.
Autonomous Artificial Intelligence Agents for Clinical Decision Making in Oncology
Apr 06, 2024

Abstract:Multimodal artificial intelligence (AI) systems have the potential to enhance clinical decision-making by interpreting various types of medical data. However, the effectiveness of these models across all medical fields is uncertain. Each discipline presents unique challenges that need to be addressed for optimal performance. This complexity is further increased when attempting to integrate different fields into a single model. Here, we introduce an alternative approach to multimodal medical AI that utilizes the generalist capabilities of a large language model (LLM) as a central reasoning engine. This engine autonomously coordinates and deploys a set of specialized medical AI tools. These tools include text, radiology and histopathology image interpretation, genomic data processing, web searches, and document retrieval from medical guidelines. We validate our system across a series of clinical oncology scenarios that closely resemble typical patient care workflows. We show that the system has a high capability in employing appropriate tools (97%), drawing correct conclusions (93.6%), and providing complete (94%), and helpful (89.2%) recommendations for individual patient cases while consistently referencing relevant literature (82.5%) upon instruction. This work provides evidence that LLMs can effectively plan and execute domain-specific models to retrieve or synthesize new information when used as autonomous agents. This enables them to function as specialist, patient-tailored clinical assistants. It also simplifies regulatory compliance by allowing each component tool to be individually validated and approved. We believe, that our work can serve as a proof-of-concept for more advanced LLM-agents in the medical domain.
Reducing self-supervised learning complexity improves weakly-supervised classification performance in computational pathology
Mar 12, 2024Abstract:Deep Learning models have been successfully utilized to extract clinically actionable insights from routinely available histology data. Generally, these models require annotations performed by clinicians, which are scarce and costly to generate. The emergence of self-supervised learning (SSL) methods remove this barrier, allowing for large-scale analyses on non-annotated data. However, recent SSL approaches apply increasingly expansive model architectures and larger datasets, causing the rapid escalation of data volumes, hardware prerequisites, and overall expenses, limiting access to these resources to few institutions. Therefore, we investigated the complexity of contrastive SSL in computational pathology in relation to classification performance with the utilization of consumer-grade hardware. Specifically, we analyzed the effects of adaptations in data volume, architecture, and algorithms on downstream classification tasks, emphasizing their impact on computational resources. We trained breast cancer foundation models on a large public patient cohort and validated them on various downstream classification tasks in a weakly supervised manner on two external public patient cohorts. Our experiments demonstrate that we can improve downstream classification performance whilst reducing SSL training duration by 90%. In summary, we propose a set of adaptations which enable the utilization of SSL in computational pathology in non-resource abundant environments.
In-context learning enables multimodal large language models to classify cancer pathology images
Mar 12, 2024Abstract:Medical image classification requires labeled, task-specific datasets which are used to train deep learning networks de novo, or to fine-tune foundation models. However, this process is computationally and technically demanding. In language processing, in-context learning provides an alternative, where models learn from within prompts, bypassing the need for parameter updates. Yet, in-context learning remains underexplored in medical image analysis. Here, we systematically evaluate the model Generative Pretrained Transformer 4 with Vision capabilities (GPT-4V) on cancer image processing with in-context learning on three cancer histopathology tasks of high importance: Classification of tissue subtypes in colorectal cancer, colon polyp subtyping and breast tumor detection in lymph node sections. Our results show that in-context learning is sufficient to match or even outperform specialized neural networks trained for particular tasks, while only requiring a minimal number of samples. In summary, this study demonstrates that large vision language models trained on non-domain specific data can be applied out-of-the box to solve medical image-processing tasks in histopathology. This democratizes access of generalist AI models to medical experts without technical background especially for areas where annotated data is scarce.
Joint multi-task learning improves weakly-supervised biomarker prediction in computational pathology
Mar 06, 2024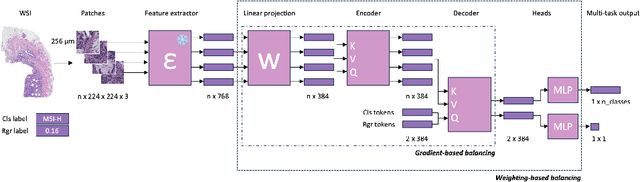


Abstract:Deep Learning (DL) can predict biomarkers directly from digitized cancer histology in a weakly-supervised setting. Recently, the prediction of continuous biomarkers through regression-based DL has seen an increasing interest. Nonetheless, clinical decision making often requires a categorical outcome. Consequently, we developed a weakly-supervised joint multi-task Transformer architecture which has been trained and evaluated on four public patient cohorts for the prediction of two key predictive biomarkers, microsatellite instability (MSI) and homologous recombination deficiency (HRD), trained with auxiliary regression tasks related to the tumor microenvironment. Moreover, we perform a comprehensive benchmark of 16 approaches of task balancing for weakly-supervised joint multi-task learning in computational pathology. Using our novel approach, we improve over the state-of-the-art area under the receiver operating characteristic by +7.7% and +4.1%, as well as yielding better clustering of latent embeddings by +8% and +5% for the prediction of MSI and HRD in external cohorts, respectively.
From Whole-slide Image to Biomarker Prediction: A Protocol for End-to-End Deep Learning in Computational Pathology
Dec 18, 2023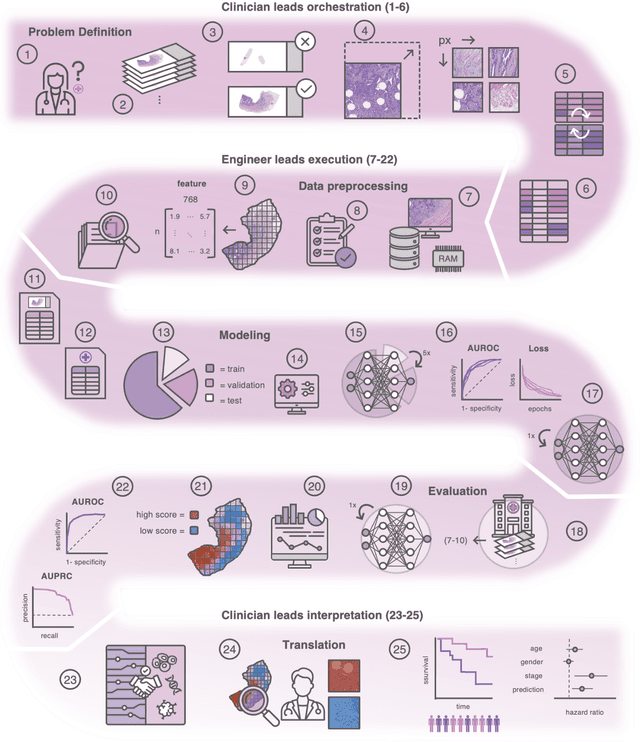
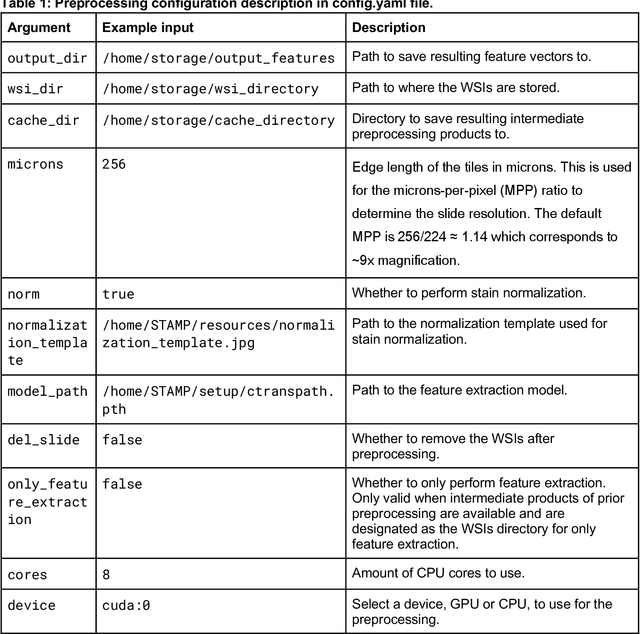
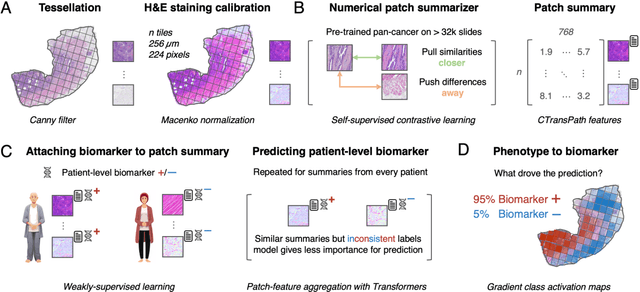

Abstract:Hematoxylin- and eosin (H&E) stained whole-slide images (WSIs) are the foundation of diagnosis of cancer. In recent years, development of deep learning-based methods in computational pathology enabled the prediction of biomarkers directly from WSIs. However, accurately linking tissue phenotype to biomarkers at scale remains a crucial challenge for democratizing complex biomarkers in precision oncology. This protocol describes a practical workflow for solid tumor associative modeling in pathology (STAMP), enabling prediction of biomarkers directly from WSIs using deep learning. The STAMP workflow is biomarker agnostic and allows for genetic- and clinicopathologic tabular data to be included as an additional input, together with histopathology images. The protocol consists of five main stages which have been successfully applied to various research problems: formal problem definition, data preprocessing, modeling, evaluation and clinical translation. The STAMP workflow differentiates itself through its focus on serving as a collaborative framework that can be used by clinicians and engineers alike for setting up research projects in the field of computational pathology. As an example task, we applied STAMP to the prediction of microsatellite instability (MSI) status in colorectal cancer, showing accurate performance for the identification of MSI-high tumors. Moreover, we provide an open-source codebase which has been deployed at several hospitals across the globe to set up computational pathology workflows. The STAMP workflow requires one workday of hands-on computational execution and basic command line knowledge.
A Good Feature Extractor Is All You Need for Weakly Supervised Learning in Histopathology
Nov 29, 2023Abstract:Deep learning is revolutionising pathology, offering novel opportunities in disease prognosis and personalised treatment. Historically, stain normalisation has been a crucial preprocessing step in computational pathology pipelines, and persists into the deep learning era. Yet, with the emergence of feature extractors trained using self-supervised learning (SSL) on diverse pathology datasets, we call this practice into question. In an empirical evaluation of publicly available feature extractors, we find that omitting stain normalisation and image augmentations does not compromise downstream performance, while incurring substantial savings in memory and compute. Further, we show that the top-performing feature extractors are remarkably robust to variations in stain and augmentations like rotation in their latent space. Contrary to previous patch-level benchmarking studies, our approach emphasises clinical relevance by focusing on slide-level prediction tasks in a weakly supervised setting with external validation cohorts. This work represents the most comprehensive robustness evaluation of public pathology SSL feature extractors to date, involving more than 6,000 training runs across nine tasks, five datasets, three downstream architectures, and various preprocessing setups. Our findings stand to streamline digital pathology workflows by minimising preprocessing needs and informing the selection of feature extractors.
Regression-based Deep-Learning predicts molecular biomarkers from pathology slides
Apr 11, 2023Abstract:Deep Learning (DL) can predict biomarkers from cancer histopathology. Several clinically approved applications use this technology. Most approaches, however, predict categorical labels, whereas biomarkers are often continuous measurements. We hypothesized that regression-based DL outperforms classification-based DL. Therefore, we developed and evaluated a new self-supervised attention-based weakly supervised regression method that predicts continuous biomarkers directly from images in 11,671 patients across nine cancer types. We tested our method for multiple clinically and biologically relevant biomarkers: homologous repair deficiency (HRD) score, a clinically used pan-cancer biomarker, as well as markers of key biological processes in the tumor microenvironment. Using regression significantly enhances the accuracy of biomarker prediction, while also improving the interpretability of the results over classification. In a large cohort of colorectal cancer patients, regression-based prediction scores provide a higher prognostic value than classification-based scores. Our open-source regression approach offers a promising alternative for continuous biomarker analysis in computational pathology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge