Sebastian Foersch
A deep learning framework for efficient pathology image analysis
Feb 18, 2025



Abstract:Artificial intelligence (AI) has transformed digital pathology by enabling biomarker prediction from high-resolution whole slide images (WSIs). However, current methods are computationally inefficient, processing thousands of redundant tiles per WSI and requiring complex aggregator models. We introduce EAGLE (Efficient Approach for Guided Local Examination), a deep learning framework that emulates pathologists by selectively analyzing informative regions. EAGLE incorporates two foundation models: CHIEF for efficient tile selection and Virchow2 for extracting high-quality features. Benchmarking was conducted against leading slide- and tile-level foundation models across 31 tasks from four cancer types, spanning morphology, biomarker prediction and prognosis. EAGLE outperformed state-of-the-art foundation models by up to 23% and achieved the highest AUROC overall. It processed a slide in 2.27 seconds, reducing computational time by more than 99% compared to existing models. This efficiency enables real-time workflows, allows pathologists to validate all tiles which are used by the model during analysis, and eliminates dependence on high-performance computing, making AI-powered pathology more accessible. By reliably identifying meaningful regions and minimizing artifacts, EAGLE provides robust and interpretable outputs, supporting rapid slide searches, integration into multi-omics pipelines and emerging clinical foundation models.
Benchmarking foundation models as feature extractors for weakly-supervised computational pathology
Aug 28, 2024Abstract:Advancements in artificial intelligence have driven the development of numerous pathology foundation models capable of extracting clinically relevant information. However, there is currently limited literature independently evaluating these foundation models on truly external cohorts and clinically-relevant tasks to uncover adjustments for future improvements. In this study, we benchmarked ten histopathology foundation models on 13 patient cohorts with 6,791 patients and 9,493 slides from lung, colorectal, gastric, and breast cancers. The models were evaluated on weakly-supervised tasks related to biomarkers, morphological properties, and prognostic outcomes. We show that a vision-language foundation model, CONCH, yielded the highest performance in 42% of tasks when compared to vision-only foundation models. The experiments reveal that foundation models trained on distinct cohorts learn complementary features to predict the same label, and can be fused to outperform the current state of the art. Creating an ensemble of complementary foundation models outperformed CONCH in 66% of tasks. Moreover, our findings suggest that data diversity outweighs data volume for foundation models. Our work highlights actionable adjustments to improve pathology foundation models.
Prompt Injection Attacks on Large Language Models in Oncology
Jul 23, 2024



Abstract:Vision-language artificial intelligence models (VLMs) possess medical knowledge and can be employed in healthcare in numerous ways, including as image interpreters, virtual scribes, and general decision support systems. However, here, we demonstrate that current VLMs applied to medical tasks exhibit a fundamental security flaw: they can be attacked by prompt injection attacks, which can be used to output harmful information just by interacting with the VLM, without any access to its parameters. We performed a quantitative study to evaluate the vulnerabilities to these attacks in four state of the art VLMs which have been proposed to be of utility in healthcare: Claude 3 Opus, Claude 3.5 Sonnet, Reka Core, and GPT-4o. Using a set of N=297 attacks, we show that all of these models are susceptible. Specifically, we show that embedding sub-visual prompts in medical imaging data can cause the model to provide harmful output, and that these prompts are non-obvious to human observers. Thus, our study demonstrates a key vulnerability in medical VLMs which should be mitigated before widespread clinical adoption.
Autonomous Artificial Intelligence Agents for Clinical Decision Making in Oncology
Apr 06, 2024

Abstract:Multimodal artificial intelligence (AI) systems have the potential to enhance clinical decision-making by interpreting various types of medical data. However, the effectiveness of these models across all medical fields is uncertain. Each discipline presents unique challenges that need to be addressed for optimal performance. This complexity is further increased when attempting to integrate different fields into a single model. Here, we introduce an alternative approach to multimodal medical AI that utilizes the generalist capabilities of a large language model (LLM) as a central reasoning engine. This engine autonomously coordinates and deploys a set of specialized medical AI tools. These tools include text, radiology and histopathology image interpretation, genomic data processing, web searches, and document retrieval from medical guidelines. We validate our system across a series of clinical oncology scenarios that closely resemble typical patient care workflows. We show that the system has a high capability in employing appropriate tools (97%), drawing correct conclusions (93.6%), and providing complete (94%), and helpful (89.2%) recommendations for individual patient cases while consistently referencing relevant literature (82.5%) upon instruction. This work provides evidence that LLMs can effectively plan and execute domain-specific models to retrieve or synthesize new information when used as autonomous agents. This enables them to function as specialist, patient-tailored clinical assistants. It also simplifies regulatory compliance by allowing each component tool to be individually validated and approved. We believe, that our work can serve as a proof-of-concept for more advanced LLM-agents in the medical domain.
From Whole-slide Image to Biomarker Prediction: A Protocol for End-to-End Deep Learning in Computational Pathology
Dec 18, 2023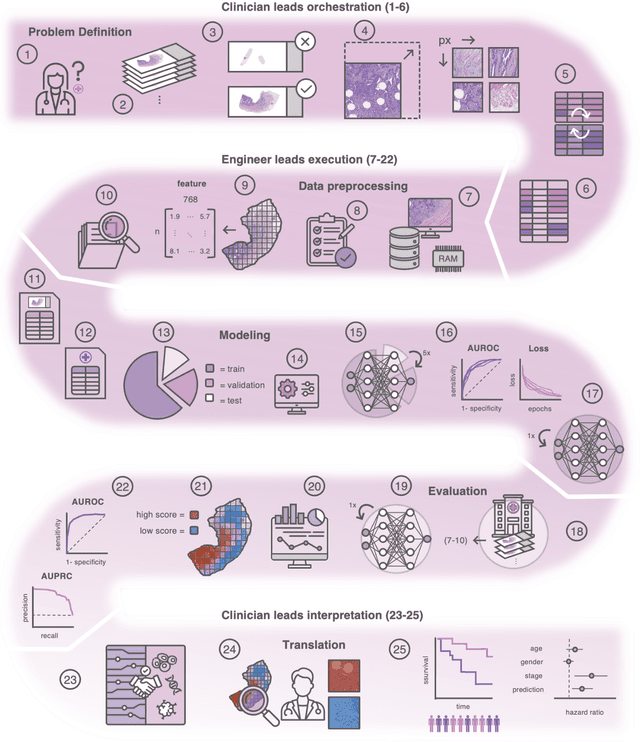
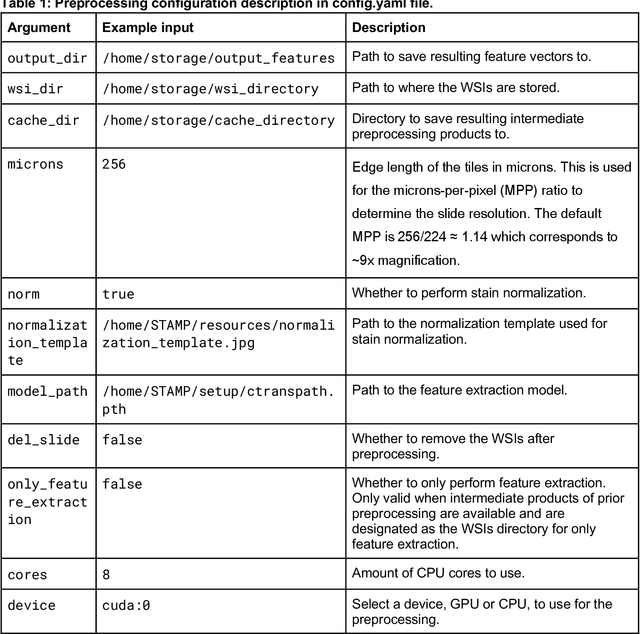
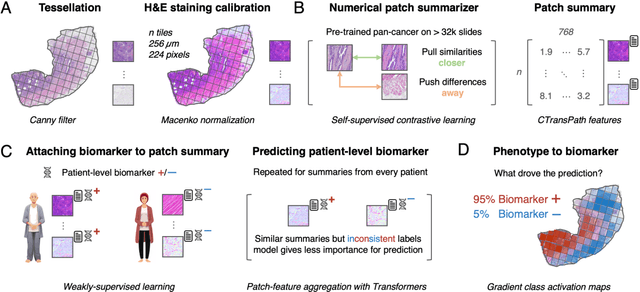

Abstract:Hematoxylin- and eosin (H&E) stained whole-slide images (WSIs) are the foundation of diagnosis of cancer. In recent years, development of deep learning-based methods in computational pathology enabled the prediction of biomarkers directly from WSIs. However, accurately linking tissue phenotype to biomarkers at scale remains a crucial challenge for democratizing complex biomarkers in precision oncology. This protocol describes a practical workflow for solid tumor associative modeling in pathology (STAMP), enabling prediction of biomarkers directly from WSIs using deep learning. The STAMP workflow is biomarker agnostic and allows for genetic- and clinicopathologic tabular data to be included as an additional input, together with histopathology images. The protocol consists of five main stages which have been successfully applied to various research problems: formal problem definition, data preprocessing, modeling, evaluation and clinical translation. The STAMP workflow differentiates itself through its focus on serving as a collaborative framework that can be used by clinicians and engineers alike for setting up research projects in the field of computational pathology. As an example task, we applied STAMP to the prediction of microsatellite instability (MSI) status in colorectal cancer, showing accurate performance for the identification of MSI-high tumors. Moreover, we provide an open-source codebase which has been deployed at several hospitals across the globe to set up computational pathology workflows. The STAMP workflow requires one workday of hands-on computational execution and basic command line knowledge.
Medical Diffusion -- Denoising Diffusion Probabilistic Models for 3D Medical Image Generation
Nov 07, 2022Abstract:Recent advances in computer vision have shown promising results in image generation. Diffusion probabilistic models in particular have generated realistic images from textual input, as demonstrated by DALL-E 2, Imagen and Stable Diffusion. However, their use in medicine, where image data typically comprises three-dimensional volumes, has not been systematically evaluated. Synthetic images may play a crucial role in privacy preserving artificial intelligence and can also be used to augment small datasets. Here we show that diffusion probabilistic models can synthesize high quality medical imaging data, which we show for Magnetic Resonance Images (MRI) and Computed Tomography (CT) images. We provide quantitative measurements of their performance through a reader study with two medical experts who rated the quality of the synthesized images in three categories: Realistic image appearance, anatomical correctness and consistency between slices. Furthermore, we demonstrate that synthetic images can be used in a self-supervised pre-training and improve the performance of breast segmentation models when data is scarce (dice score 0.91 vs. 0.95 without vs. with synthetic data).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge