Jacqueline Lammert
Large Language Models-Enabled Digital Twins for Precision Medicine in Rare Gynecological Tumors
Aug 31, 2024Abstract:Rare gynecological tumors (RGTs) present major clinical challenges due to their low incidence and heterogeneity. The lack of clear guidelines leads to suboptimal management and poor prognosis. Molecular tumor boards accelerate access to effective therapies by tailoring treatment based on biomarkers, beyond cancer type. Unstructured data that requires manual curation hinders efficient use of biomarker profiling for therapy matching. This study explores the use of large language models (LLMs) to construct digital twins for precision medicine in RGTs. Our proof-of-concept digital twin system integrates clinical and biomarker data from institutional and published cases (n=21) and literature-derived data (n=655 publications with n=404,265 patients) to create tailored treatment plans for metastatic uterine carcinosarcoma, identifying options potentially missed by traditional, single-source analysis. LLM-enabled digital twins efficiently model individual patient trajectories. Shifting to a biology-based rather than organ-based tumor definition enables personalized care that could advance RGT management and thus enhance patient outcomes.
Biomedical Large Languages Models Seem not to be Superior to Generalist Models on Unseen Medical Data
Aug 25, 2024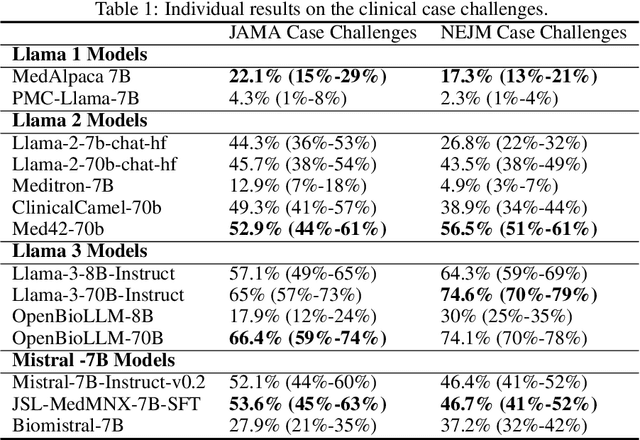
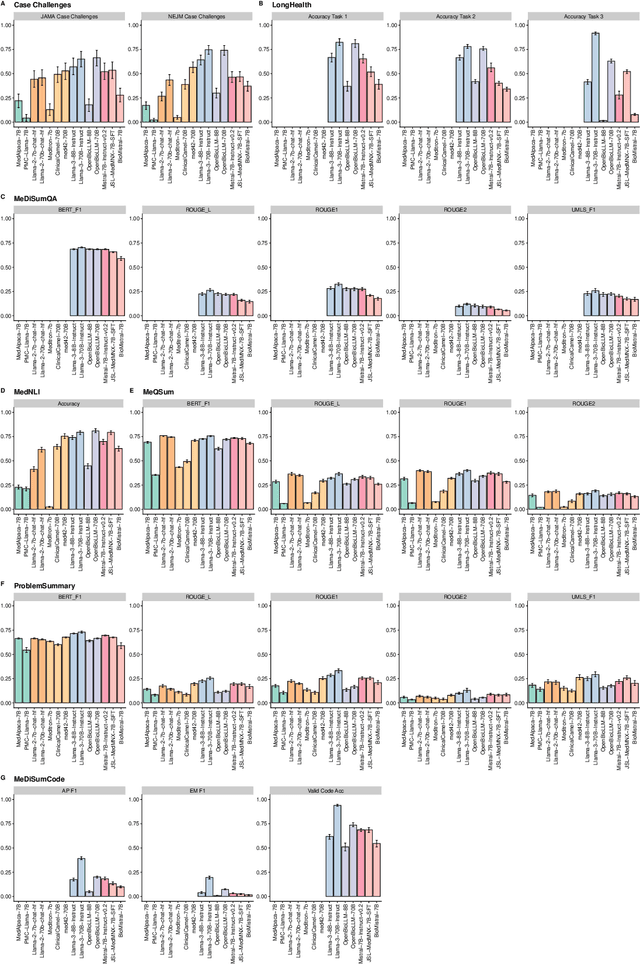
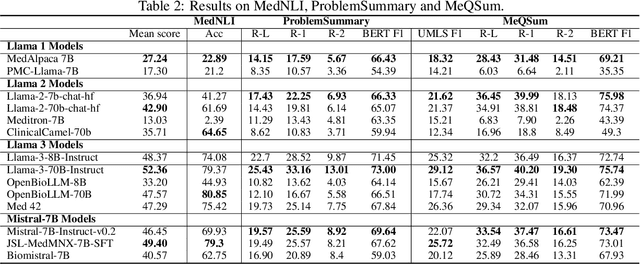
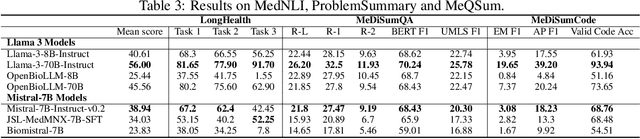
Abstract:Large language models (LLMs) have shown potential in biomedical applications, leading to efforts to fine-tune them on domain-specific data. However, the effectiveness of this approach remains unclear. This study evaluates the performance of biomedically fine-tuned LLMs against their general-purpose counterparts on a variety of clinical tasks. We evaluated their performance on clinical case challenges from the New England Journal of Medicine (NEJM) and the Journal of the American Medical Association (JAMA) and on several clinical tasks (e.g., information extraction, document summarization, and clinical coding). Using benchmarks specifically chosen to be likely outside the fine-tuning datasets of biomedical models, we found that biomedical LLMs mostly perform inferior to their general-purpose counterparts, especially on tasks not focused on medical knowledge. While larger models showed similar performance on case tasks (e.g., OpenBioLLM-70B: 66.4% vs. Llama-3-70B-Instruct: 65% on JAMA cases), smaller biomedical models showed more pronounced underperformance (e.g., OpenBioLLM-8B: 30% vs. Llama-3-8B-Instruct: 64.3% on NEJM cases). Similar trends were observed across the CLUE (Clinical Language Understanding Evaluation) benchmark tasks, with general-purpose models often performing better on text generation, question answering, and coding tasks. Our results suggest that fine-tuning LLMs to biomedical data may not provide the expected benefits and may potentially lead to reduced performance, challenging prevailing assumptions about domain-specific adaptation of LLMs and highlighting the need for more rigorous evaluation frameworks in healthcare AI. Alternative approaches, such as retrieval-augmented generation, may be more effective in enhancing the biomedical capabilities of LLMs without compromising their general knowledge.
End-To-End Clinical Trial Matching with Large Language Models
Jul 18, 2024


Abstract:Matching cancer patients to clinical trials is essential for advancing treatment and patient care. However, the inconsistent format of medical free text documents and complex trial eligibility criteria make this process extremely challenging and time-consuming for physicians. We investigated whether the entire trial matching process - from identifying relevant trials among 105,600 oncology-related clinical trials on clinicaltrials.gov to generating criterion-level eligibility matches - could be automated using Large Language Models (LLMs). Using GPT-4o and a set of 51 synthetic Electronic Health Records (EHRs), we demonstrate that our approach identifies relevant candidate trials in 93.3% of cases and achieves a preliminary accuracy of 88.0% when matching patient-level information at the criterion level against a baseline defined by human experts. Utilizing LLM feedback reveals that 39.3% criteria that were initially considered incorrect are either ambiguous or inaccurately annotated, leading to a total model accuracy of 92.7% after refining our human baseline. In summary, we present an end-to-end pipeline for clinical trial matching using LLMs, demonstrating high precision in screening and matching trials to individual patients, even outperforming the performance of qualified medical doctors. Our fully end-to-end pipeline can operate autonomously or with human supervision and is not restricted to oncology, offering a scalable solution for enhancing patient-trial matching in real-world settings.
Autonomous Artificial Intelligence Agents for Clinical Decision Making in Oncology
Apr 06, 2024

Abstract:Multimodal artificial intelligence (AI) systems have the potential to enhance clinical decision-making by interpreting various types of medical data. However, the effectiveness of these models across all medical fields is uncertain. Each discipline presents unique challenges that need to be addressed for optimal performance. This complexity is further increased when attempting to integrate different fields into a single model. Here, we introduce an alternative approach to multimodal medical AI that utilizes the generalist capabilities of a large language model (LLM) as a central reasoning engine. This engine autonomously coordinates and deploys a set of specialized medical AI tools. These tools include text, radiology and histopathology image interpretation, genomic data processing, web searches, and document retrieval from medical guidelines. We validate our system across a series of clinical oncology scenarios that closely resemble typical patient care workflows. We show that the system has a high capability in employing appropriate tools (97%), drawing correct conclusions (93.6%), and providing complete (94%), and helpful (89.2%) recommendations for individual patient cases while consistently referencing relevant literature (82.5%) upon instruction. This work provides evidence that LLMs can effectively plan and execute domain-specific models to retrieve or synthesize new information when used as autonomous agents. This enables them to function as specialist, patient-tailored clinical assistants. It also simplifies regulatory compliance by allowing each component tool to be individually validated and approved. We believe, that our work can serve as a proof-of-concept for more advanced LLM-agents in the medical domain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge