Felix Busch
Improving Reliability and Explainability of Medical Question Answering through Atomic Fact Checking in Retrieval-Augmented LLMs
May 30, 2025Abstract:Large language models (LLMs) exhibit extensive medical knowledge but are prone to hallucinations and inaccurate citations, which pose a challenge to their clinical adoption and regulatory compliance. Current methods, such as Retrieval Augmented Generation, partially address these issues by grounding answers in source documents, but hallucinations and low fact-level explainability persist. In this work, we introduce a novel atomic fact-checking framework designed to enhance the reliability and explainability of LLMs used in medical long-form question answering. This method decomposes LLM-generated responses into discrete, verifiable units called atomic facts, each of which is independently verified against an authoritative knowledge base of medical guidelines. This approach enables targeted correction of errors and direct tracing to source literature, thereby improving the factual accuracy and explainability of medical Q&A. Extensive evaluation using multi-reader assessments by medical experts and an automated open Q&A benchmark demonstrated significant improvements in factual accuracy and explainability. Our framework achieved up to a 40% overall answer improvement and a 50% hallucination detection rate. The ability to trace each atomic fact back to the most relevant chunks from the database provides a granular, transparent explanation of the generated responses, addressing a major gap in current medical AI applications. This work represents a crucial step towards more trustworthy and reliable clinical applications of LLMs, addressing key prerequisites for clinical application and fostering greater confidence in AI-assisted healthcare.
Biomedical Large Languages Models Seem not to be Superior to Generalist Models on Unseen Medical Data
Aug 25, 2024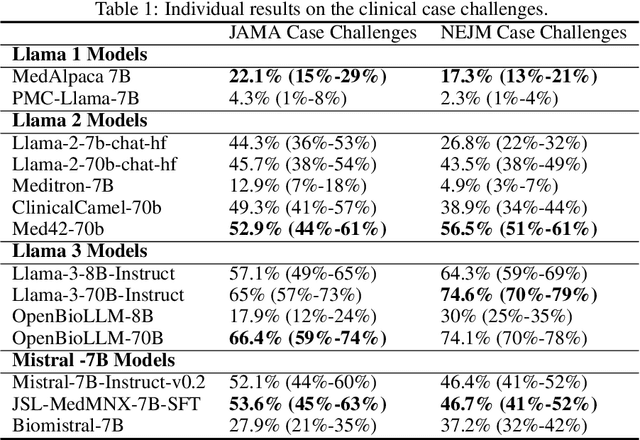
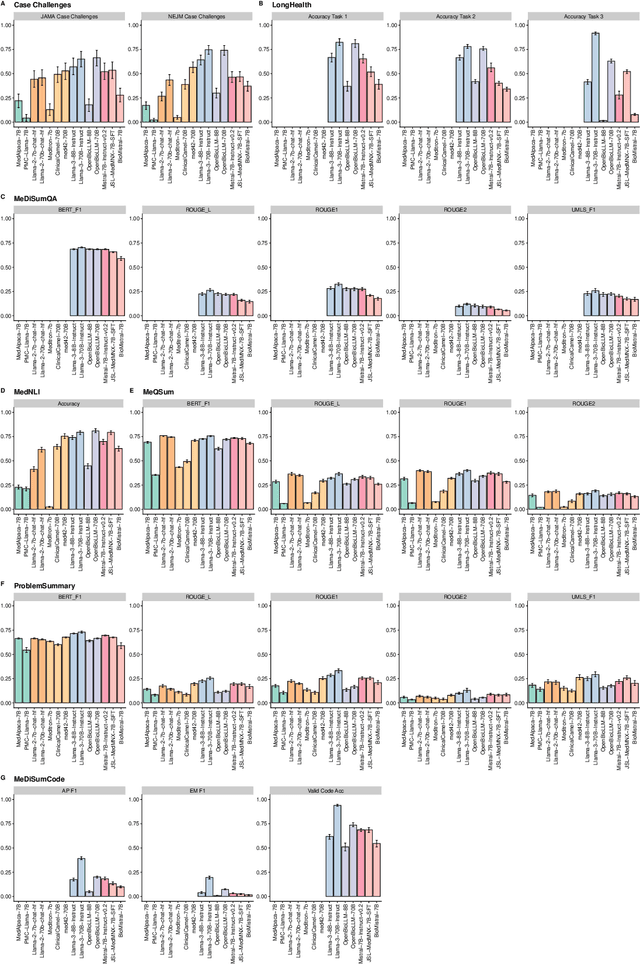
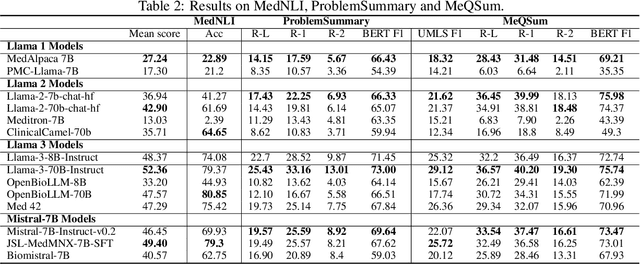
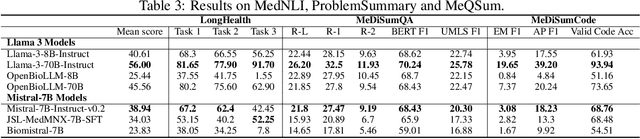
Abstract:Large language models (LLMs) have shown potential in biomedical applications, leading to efforts to fine-tune them on domain-specific data. However, the effectiveness of this approach remains unclear. This study evaluates the performance of biomedically fine-tuned LLMs against their general-purpose counterparts on a variety of clinical tasks. We evaluated their performance on clinical case challenges from the New England Journal of Medicine (NEJM) and the Journal of the American Medical Association (JAMA) and on several clinical tasks (e.g., information extraction, document summarization, and clinical coding). Using benchmarks specifically chosen to be likely outside the fine-tuning datasets of biomedical models, we found that biomedical LLMs mostly perform inferior to their general-purpose counterparts, especially on tasks not focused on medical knowledge. While larger models showed similar performance on case tasks (e.g., OpenBioLLM-70B: 66.4% vs. Llama-3-70B-Instruct: 65% on JAMA cases), smaller biomedical models showed more pronounced underperformance (e.g., OpenBioLLM-8B: 30% vs. Llama-3-8B-Instruct: 64.3% on NEJM cases). Similar trends were observed across the CLUE (Clinical Language Understanding Evaluation) benchmark tasks, with general-purpose models often performing better on text generation, question answering, and coding tasks. Our results suggest that fine-tuning LLMs to biomedical data may not provide the expected benefits and may potentially lead to reduced performance, challenging prevailing assumptions about domain-specific adaptation of LLMs and highlighting the need for more rigorous evaluation frameworks in healthcare AI. Alternative approaches, such as retrieval-augmented generation, may be more effective in enhancing the biomedical capabilities of LLMs without compromising their general knowledge.
Is Open-Source There Yet? A Comparative Study on Commercial and Open-Source LLMs in Their Ability to Label Chest X-Ray Reports
Feb 19, 2024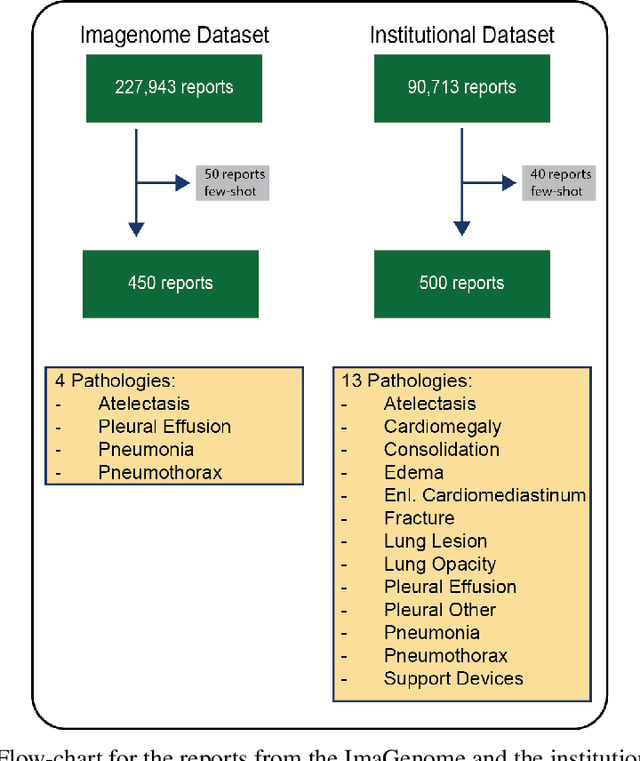
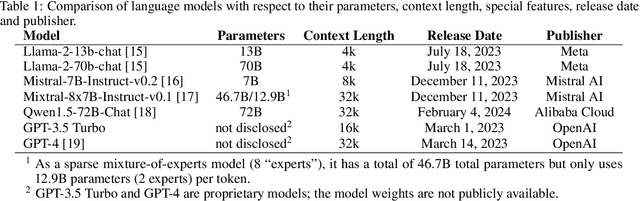
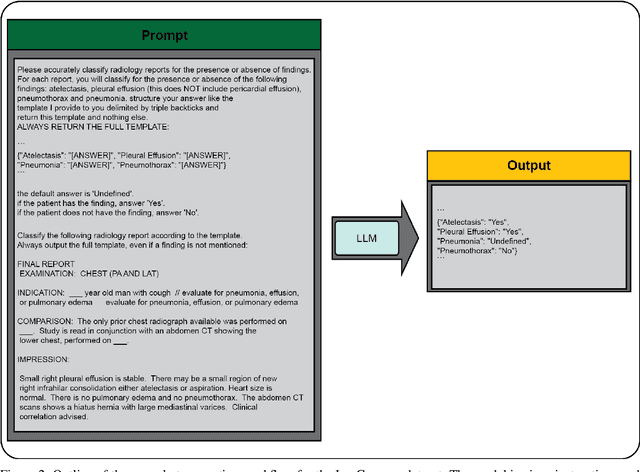
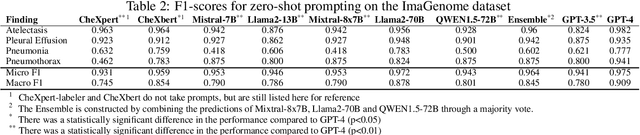
Abstract:Introduction: With the rapid advances in large language models (LLMs), there have been numerous new open source as well as commercial models. While recent publications have explored GPT-4 in its application to extracting information of interest from radiology reports, there has not been a real-world comparison of GPT-4 to different leading open-source models. Materials and Methods: Two different and independent datasets were used. The first dataset consists of 540 chest x-ray reports that were created at the Massachusetts General Hospital between July 2019 and July 2021. The second dataset consists of 500 chest x-ray reports from the ImaGenome dataset. We then compared the commercial models GPT-3.5 Turbo and GPT-4 from OpenAI to the open-source models Mistral-7B, Mixtral-8x7B, Llama2-13B, Llama2-70B, QWEN1.5-72B and CheXbert and CheXpert-labeler in their ability to accurately label the presence of multiple findings in x-ray text reports using different prompting techniques. Results: On the ImaGenome dataset, the best performing open-source model was Llama2-70B with micro F1-scores of 0.972 and 0.970 for zero- and few-shot prompts, respectively. GPT-4 achieved micro F1-scores of 0.975 and 0.984, respectively. On the institutional dataset, the best performing open-source model was QWEN1.5-72B with micro F1-scores of 0.952 and 0.965 for zero- and few-shot prompting, respectively. GPT-4 achieved micro F1-scores of 0.975 and 0.973, respectively. Conclusion: In this paper, we show that while GPT-4 is superior to open-source models in zero-shot report labeling, the implementation of few-shot prompting can bring open-source models on par with GPT-4. This shows that open-source models could be a performant and privacy preserving alternative to GPT-4 for the task of radiology report classification.
LongHealth: A Question Answering Benchmark with Long Clinical Documents
Jan 25, 2024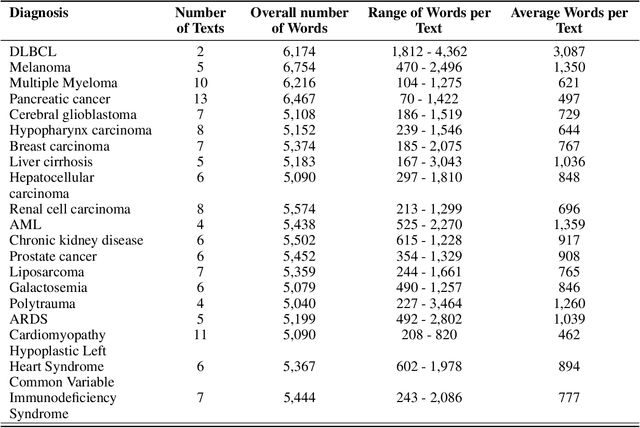
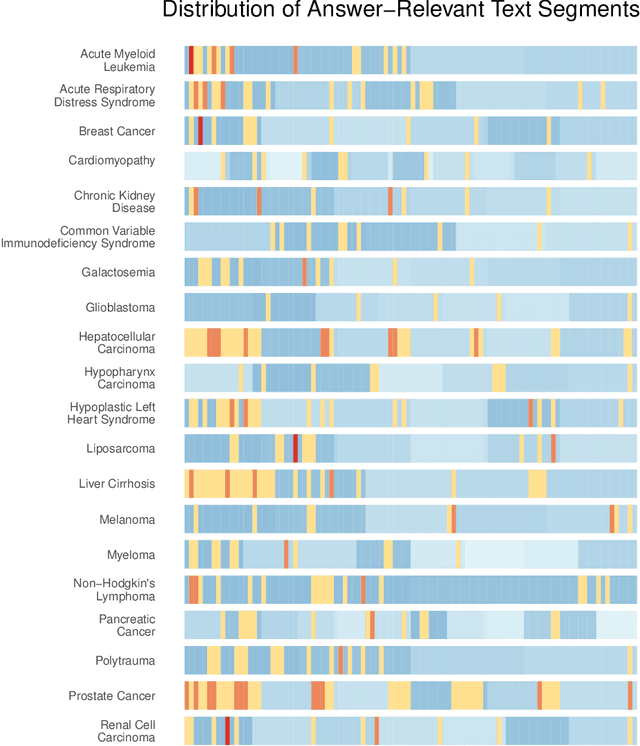

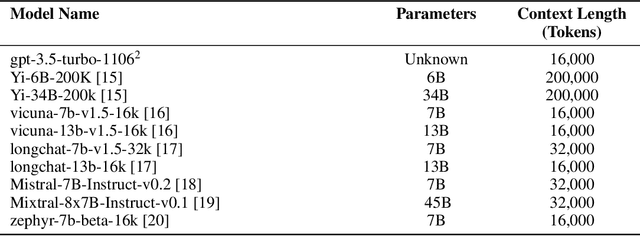
Abstract:Background: Recent advancements in large language models (LLMs) offer potential benefits in healthcare, particularly in processing extensive patient records. However, existing benchmarks do not fully assess LLMs' capability in handling real-world, lengthy clinical data. Methods: We present the LongHealth benchmark, comprising 20 detailed fictional patient cases across various diseases, with each case containing 5,090 to 6,754 words. The benchmark challenges LLMs with 400 multiple-choice questions in three categories: information extraction, negation, and sorting, challenging LLMs to extract and interpret information from large clinical documents. Results: We evaluated nine open-source LLMs with a minimum of 16,000 tokens and also included OpenAI's proprietary and cost-efficient GPT-3.5 Turbo for comparison. The highest accuracy was observed for Mixtral-8x7B-Instruct-v0.1, particularly in tasks focused on information retrieval from single and multiple patient documents. However, all models struggled significantly in tasks requiring the identification of missing information, highlighting a critical area for improvement in clinical data interpretation. Conclusion: While LLMs show considerable potential for processing long clinical documents, their current accuracy levels are insufficient for reliable clinical use, especially in scenarios requiring the identification of missing information. The LongHealth benchmark provides a more realistic assessment of LLMs in a healthcare setting and highlights the need for further model refinement for safe and effective clinical application. We make the benchmark and evaluation code publicly available.
From Text to Image: Exploring GPT-4Vision's Potential in Advanced Radiological Analysis across Subspecialties
Nov 24, 2023

Abstract:The study evaluates and compares GPT-4 and GPT-4Vision for radiological tasks, suggesting GPT-4Vision may recognize radiological features from images, thereby enhancing its diagnostic potential over text-based descriptions.
MEDBERT.de: A Comprehensive German BERT Model for the Medical Domain
Mar 24, 2023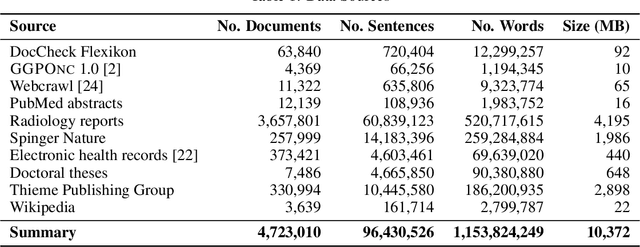
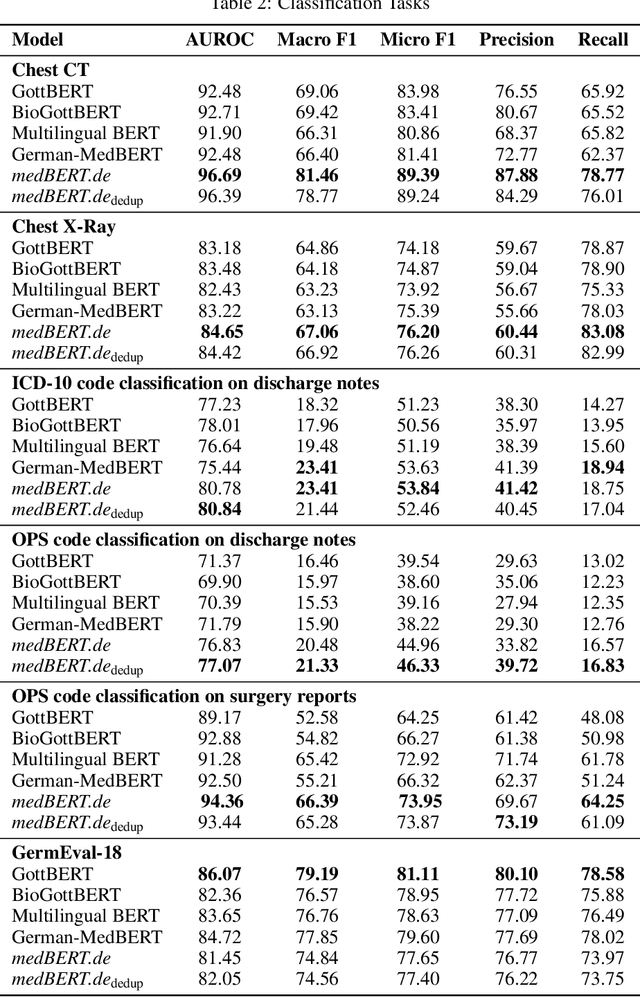
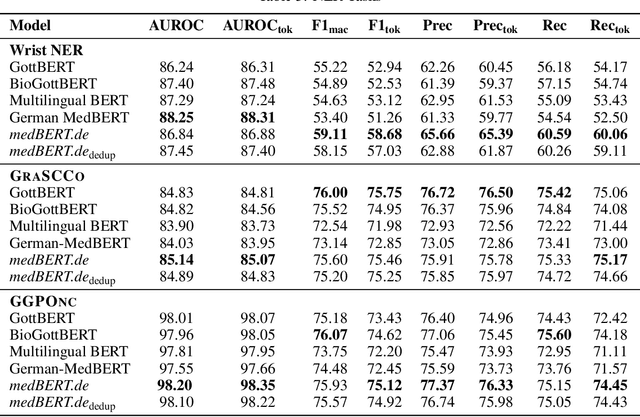
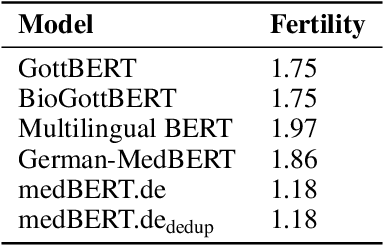
Abstract:This paper presents medBERTde, a pre-trained German BERT model specifically designed for the German medical domain. The model has been trained on a large corpus of 4.7 Million German medical documents and has been shown to achieve new state-of-the-art performance on eight different medical benchmarks covering a wide range of disciplines and medical document types. In addition to evaluating the overall performance of the model, this paper also conducts a more in-depth analysis of its capabilities. We investigate the impact of data deduplication on the model's performance, as well as the potential benefits of using more efficient tokenization methods. Our results indicate that domain-specific models such as medBERTde are particularly useful for longer texts, and that deduplication of training data does not necessarily lead to improved performance. Furthermore, we found that efficient tokenization plays only a minor role in improving model performance, and attribute most of the improved performance to the large amount of training data. To encourage further research, the pre-trained model weights and new benchmarks based on radiological data are made publicly available for use by the scientific community.
What Does DALL-E 2 Know About Radiology?
Sep 27, 2022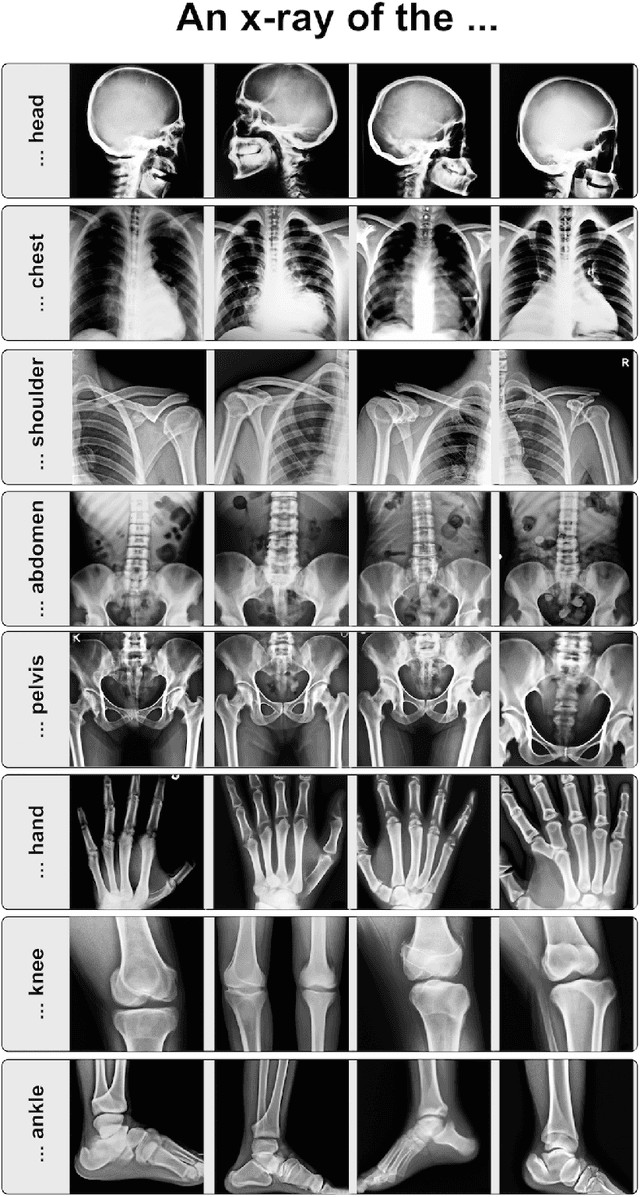
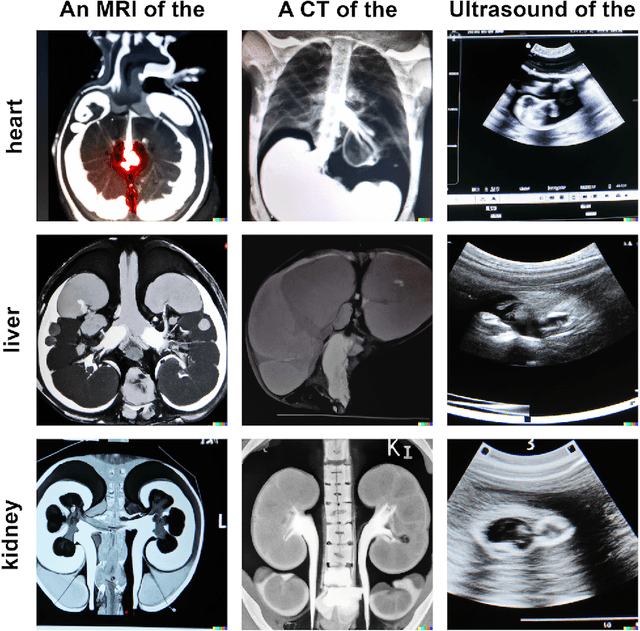
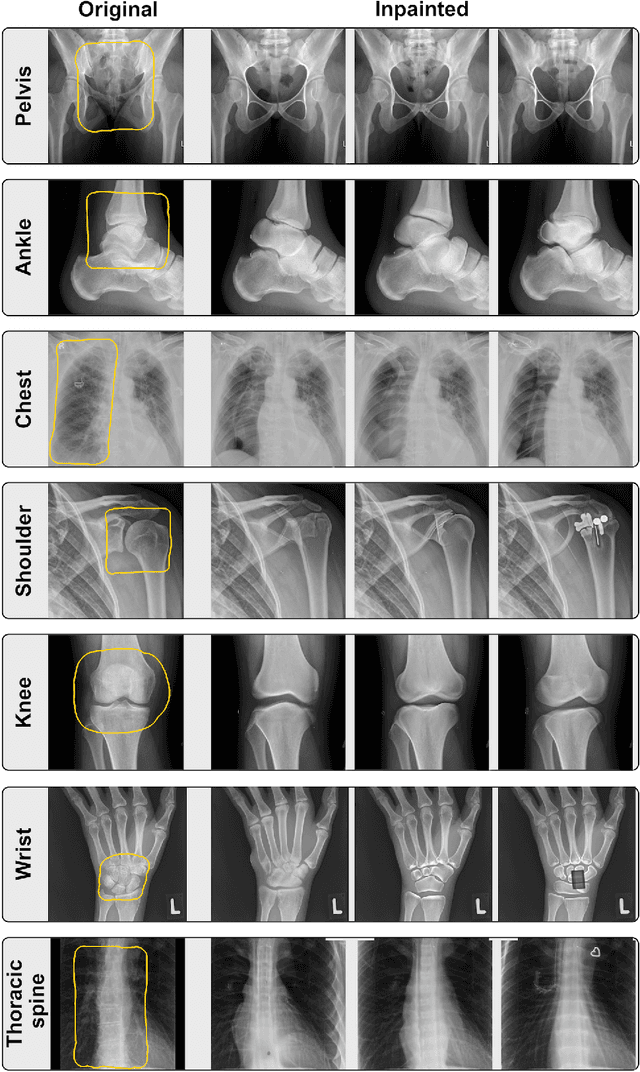
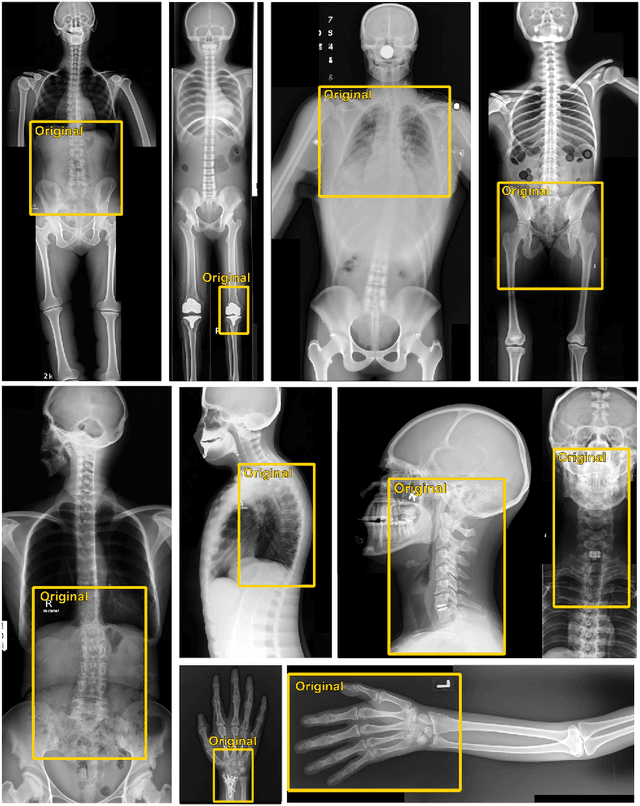
Abstract:Generative models such as DALL-E 2 could represent a promising future tool for image generation, augmentation, and manipulation for artificial intelligence research in radiology provided that these models have sufficient medical domain knowledge. Here we show that DALL-E 2 has learned relevant representations of X-ray images with promising capabilities in terms of zero-shot text-to-image generation of new images, continuation of an image beyond its original boundaries, or removal of elements, while pathology generation or CT, MRI, and ultrasound images are still limited. The use of generative models for augmenting and generating radiological data thus seems feasible, even if further fine-tuning and adaptation of these models to the respective domain is required beforehand.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge