Matthieu Ortala
LongHealth: A Question Answering Benchmark with Long Clinical Documents
Jan 25, 2024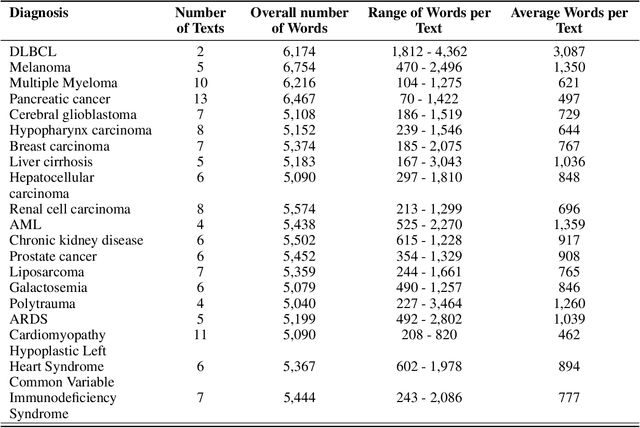
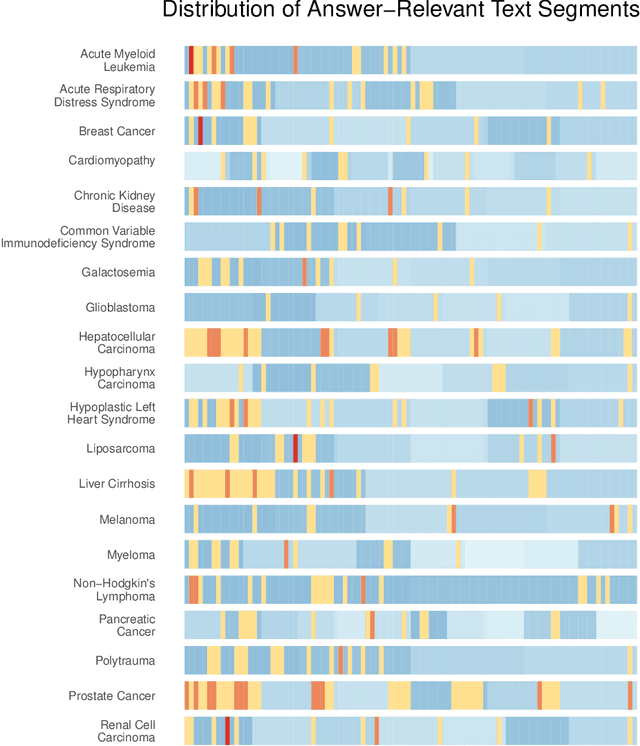

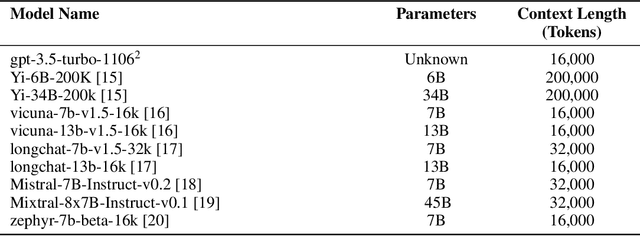
Abstract:Background: Recent advancements in large language models (LLMs) offer potential benefits in healthcare, particularly in processing extensive patient records. However, existing benchmarks do not fully assess LLMs' capability in handling real-world, lengthy clinical data. Methods: We present the LongHealth benchmark, comprising 20 detailed fictional patient cases across various diseases, with each case containing 5,090 to 6,754 words. The benchmark challenges LLMs with 400 multiple-choice questions in three categories: information extraction, negation, and sorting, challenging LLMs to extract and interpret information from large clinical documents. Results: We evaluated nine open-source LLMs with a minimum of 16,000 tokens and also included OpenAI's proprietary and cost-efficient GPT-3.5 Turbo for comparison. The highest accuracy was observed for Mixtral-8x7B-Instruct-v0.1, particularly in tasks focused on information retrieval from single and multiple patient documents. However, all models struggled significantly in tasks requiring the identification of missing information, highlighting a critical area for improvement in clinical data interpretation. Conclusion: While LLMs show considerable potential for processing long clinical documents, their current accuracy levels are insufficient for reliable clinical use, especially in scenarios requiring the identification of missing information. The LongHealth benchmark provides a more realistic assessment of LLMs in a healthcare setting and highlights the need for further model refinement for safe and effective clinical application. We make the benchmark and evaluation code publicly available.
Generalist embedding models are better at short-context clinical semantic search than specialized embedding models
Jan 06, 2024Abstract:The increasing use of tools and solutions based on Large Language Models (LLMs) for various tasks in the medical domain has become a prominent trend. Their use in this highly critical and sensitive domain has thus raised important questions about their robustness, especially in response to variations in input, and the reliability of the generated outputs. This study addresses these questions by constructing a textual dataset based on the ICD-10-CM code descriptions, widely used in US hospitals and containing many clinical terms, and their easily reproducible rephrasing. We then benchmarked existing embedding models, either generalist or specialized in the clinical domain, in a semantic search task where the goal was to correctly match the rephrased text to the original description. Our results showed that generalist models performed better than clinical models, suggesting that existing clinical specialized models are more sensitive to small changes in input that confuse them. The highlighted problem of specialized models may be due to the fact that they have not been trained on sufficient data, and in particular on datasets that are not diverse enough to have a reliable global language understanding, which is still necessary for accurate handling of medical documents.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge