Alexander Löser
"Where does it hurt?" -- Dataset and Study on Physician Intent Trajectories in Doctor Patient Dialogues
Aug 26, 2025Abstract:In a doctor-patient dialogue, the primary objective of physicians is to diagnose patients and propose a treatment plan. Medical doctors guide these conversations through targeted questioning to efficiently gather the information required to provide the best possible outcomes for patients. To the best of our knowledge, this is the first work that studies physician intent trajectories in doctor-patient dialogues. We use the `Ambient Clinical Intelligence Benchmark' (Aci-bench) dataset for our study. We collaborate with medical professionals to develop a fine-grained taxonomy of physician intents based on the SOAP framework (Subjective, Objective, Assessment, and Plan). We then conduct a large-scale annotation effort to label over 5000 doctor-patient turns with the help of a large number of medical experts recruited using Prolific, a popular crowd-sourcing platform. This large labeled dataset is an important resource contribution that we use for benchmarking the state-of-the-art generative and encoder models for medical intent classification tasks. Our findings show that our models understand the general structure of medical dialogues with high accuracy, but often fail to identify transitions between SOAP categories. We also report for the first time common trajectories in medical dialogue structures that provide valuable insights for designing `differential diagnosis' systems. Finally, we extensively study the impact of intent filtering for medical dialogue summarization and observe a significant boost in performance. We make the codes and data, including annotation guidelines, publicly available at https://github.com/DATEXIS/medical-intent-classification.
Robust Weight Imprinting: Insights from Neural Collapse and Proxy-Based Aggregation
Mar 18, 2025Abstract:The capacity of a foundation model allows for adaptation to new downstream tasks. Weight imprinting is a universal and efficient method to fulfill this purpose. It has been reinvented several times, but it has not been systematically studied. In this paper, we propose a framework for imprinting, identifying three main components: generation, normalization, and aggregation. This allows us to conduct an in-depth analysis of imprinting and a comparison of the existing work. We reveal the benefits of representing novel data with multiple proxies in the generation step and show the importance of proper normalization. We determine those proxies through clustering and propose a novel variant of imprinting that outperforms previous work. We motivate this by the neural collapse phenomenon -- an important connection that we can draw for the first time. Our results show an increase of up to 4% in challenging scenarios with complex data distributions for new classes.
Comply: Learning Sentences with Complex Weights inspired by Fruit Fly Olfaction
Feb 03, 2025Abstract:Biologically inspired neural networks offer alternative avenues to model data distributions. FlyVec is a recent example that draws inspiration from the fruit fly's olfactory circuit to tackle the task of learning word embeddings. Surprisingly, this model performs competitively even against deep learning approaches specifically designed to encode text, and it does so with the highest degree of computational efficiency. We pose the question of whether this performance can be improved further. For this, we introduce Comply. By incorporating positional information through complex weights, we enable a single-layer neural network to learn sequence representations. Our experiments show that Comply not only supersedes FlyVec but also performs on par with significantly larger state-of-the-art models. We achieve this without additional parameters. Comply yields sparse contextual representations of sentences that can be interpreted explicitly from the neuron weights.
LongHealth: A Question Answering Benchmark with Long Clinical Documents
Jan 25, 2024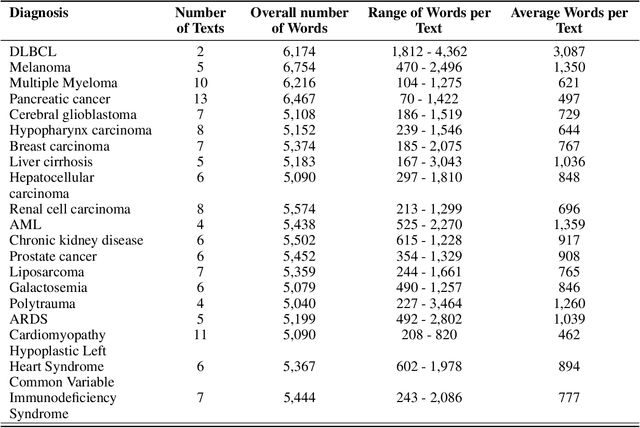
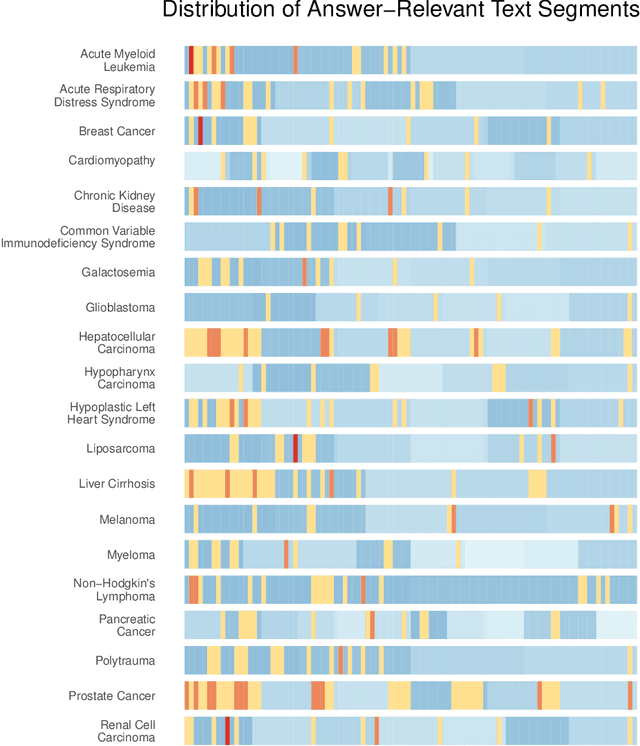

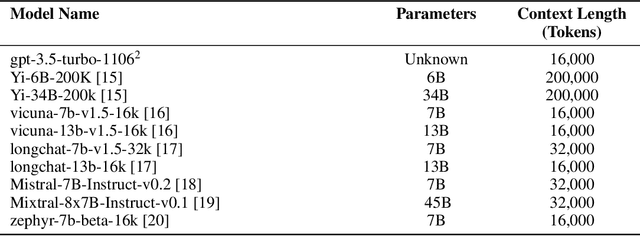
Abstract:Background: Recent advancements in large language models (LLMs) offer potential benefits in healthcare, particularly in processing extensive patient records. However, existing benchmarks do not fully assess LLMs' capability in handling real-world, lengthy clinical data. Methods: We present the LongHealth benchmark, comprising 20 detailed fictional patient cases across various diseases, with each case containing 5,090 to 6,754 words. The benchmark challenges LLMs with 400 multiple-choice questions in three categories: information extraction, negation, and sorting, challenging LLMs to extract and interpret information from large clinical documents. Results: We evaluated nine open-source LLMs with a minimum of 16,000 tokens and also included OpenAI's proprietary and cost-efficient GPT-3.5 Turbo for comparison. The highest accuracy was observed for Mixtral-8x7B-Instruct-v0.1, particularly in tasks focused on information retrieval from single and multiple patient documents. However, all models struggled significantly in tasks requiring the identification of missing information, highlighting a critical area for improvement in clinical data interpretation. Conclusion: While LLMs show considerable potential for processing long clinical documents, their current accuracy levels are insufficient for reliable clinical use, especially in scenarios requiring the identification of missing information. The LongHealth benchmark provides a more realistic assessment of LLMs in a healthcare setting and highlights the need for further model refinement for safe and effective clinical application. We make the benchmark and evaluation code publicly available.
MedAlpaca -- An Open-Source Collection of Medical Conversational AI Models and Training Data
Apr 14, 2023Abstract:As large language models (LLMs) like OpenAI's GPT series continue to make strides, we witness the emergence of artificial intelligence applications in an ever-expanding range of fields. In medicine, these LLMs hold considerable promise for improving medical workflows, diagnostics, patient care, and education. Yet, there is an urgent need for open-source models that can be deployed on-premises to safeguard patient privacy. In our work, we present an innovative dataset consisting of over 160,000 entries, specifically crafted to fine-tune LLMs for effective medical applications. We investigate the impact of fine-tuning these datasets on publicly accessible pre-trained LLMs, and subsequently, we juxtapose the performance of pre-trained-only models against the fine-tuned models concerning the examinations that future medical doctors must pass to achieve certification.
MEDBERT.de: A Comprehensive German BERT Model for the Medical Domain
Mar 24, 2023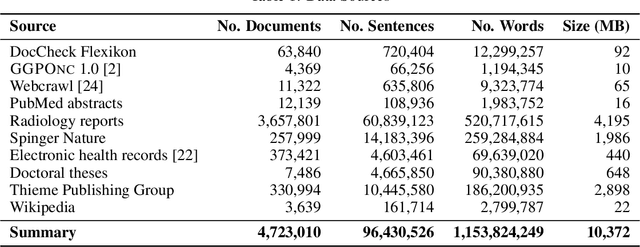
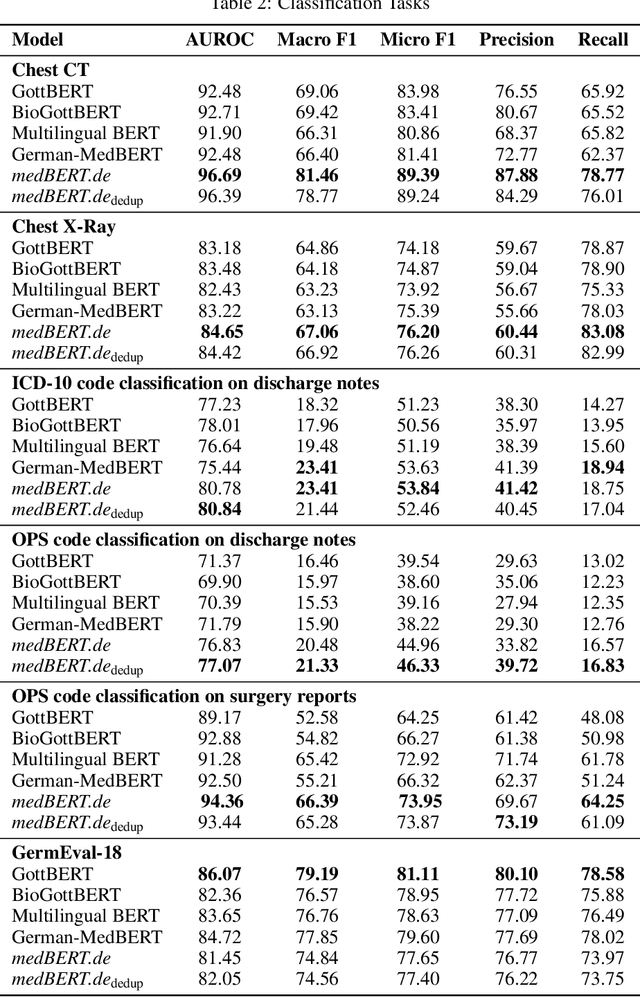
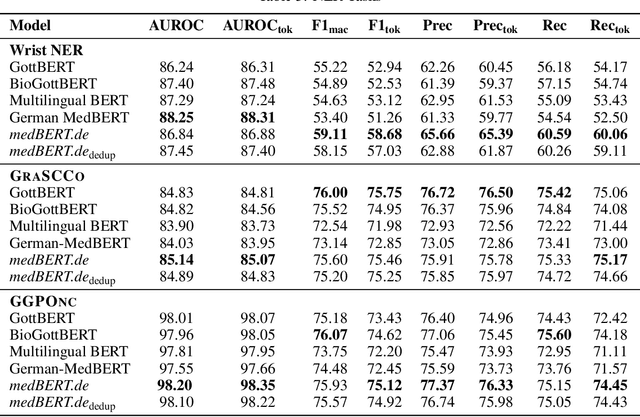
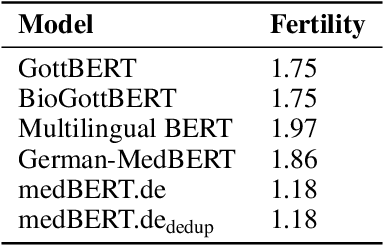
Abstract:This paper presents medBERTde, a pre-trained German BERT model specifically designed for the German medical domain. The model has been trained on a large corpus of 4.7 Million German medical documents and has been shown to achieve new state-of-the-art performance on eight different medical benchmarks covering a wide range of disciplines and medical document types. In addition to evaluating the overall performance of the model, this paper also conducts a more in-depth analysis of its capabilities. We investigate the impact of data deduplication on the model's performance, as well as the potential benefits of using more efficient tokenization methods. Our results indicate that domain-specific models such as medBERTde are particularly useful for longer texts, and that deduplication of training data does not necessarily lead to improved performance. Furthermore, we found that efficient tokenization plays only a minor role in improving model performance, and attribute most of the improved performance to the large amount of training data. To encourage further research, the pre-trained model weights and new benchmarks based on radiological data are made publicly available for use by the scientific community.
This Patient Looks Like That Patient: Prototypical Networks for Interpretable Diagnosis Prediction from Clinical Text
Oct 16, 2022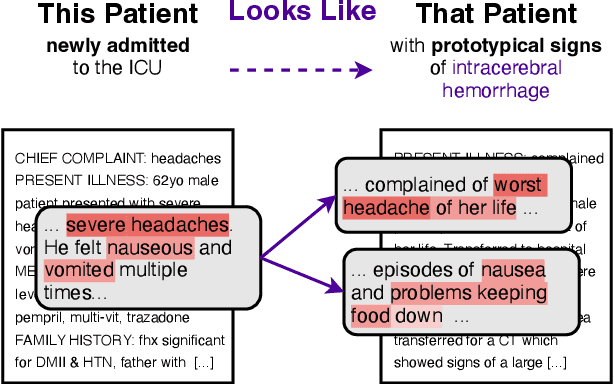
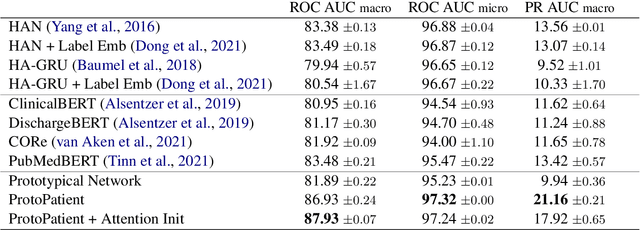
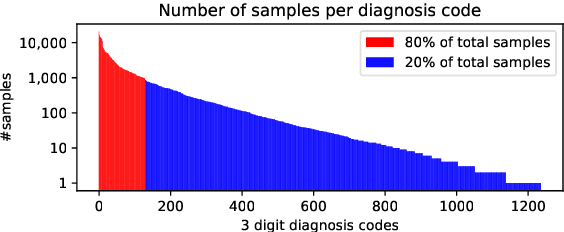
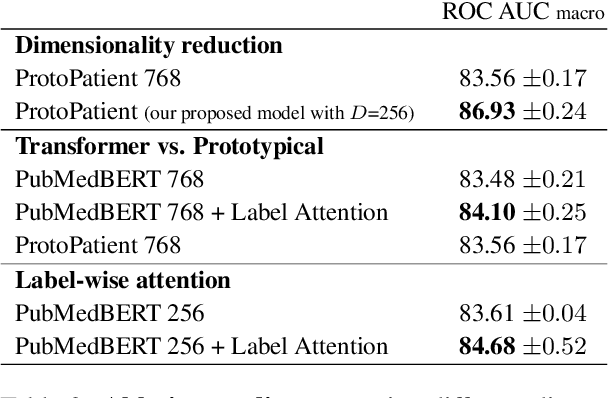
Abstract:The use of deep neural models for diagnosis prediction from clinical text has shown promising results. However, in clinical practice such models must not only be accurate, but provide doctors with interpretable and helpful results. We introduce ProtoPatient, a novel method based on prototypical networks and label-wise attention with both of these abilities. ProtoPatient makes predictions based on parts of the text that are similar to prototypical patients - providing justifications that doctors understand. We evaluate the model on two publicly available clinical datasets and show that it outperforms existing baselines. Quantitative and qualitative evaluations with medical doctors further demonstrate that the model provides valuable explanations for clinical decision support.
Cross-Lingual Knowledge Transfer for Clinical Phenotyping
Aug 03, 2022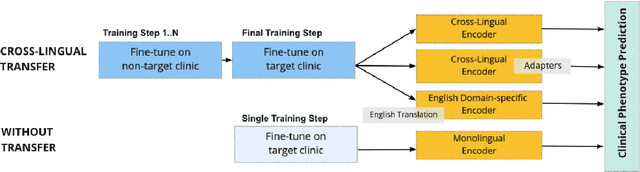
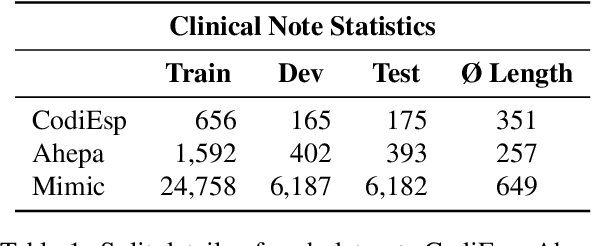

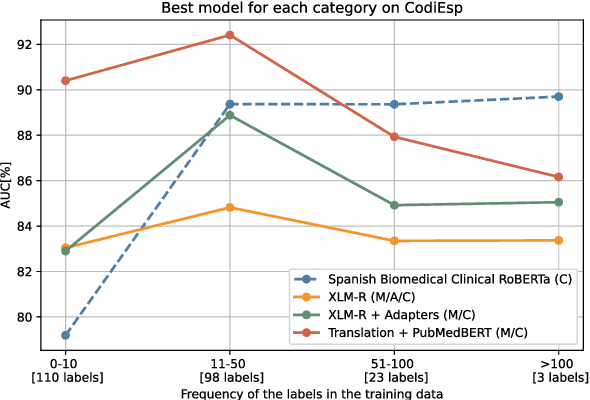
Abstract:Clinical phenotyping enables the automatic extraction of clinical conditions from patient records, which can be beneficial to doctors and clinics worldwide. However, current state-of-the-art models are mostly applicable to clinical notes written in English. We therefore investigate cross-lingual knowledge transfer strategies to execute this task for clinics that do not use the English language and have a small amount of in-domain data available. We evaluate these strategies for a Greek and a Spanish clinic leveraging clinical notes from different clinical domains such as cardiology, oncology and the ICU. Our results reveal two strategies that outperform the state-of-the-art: Translation-based methods in combination with domain-specific encoders and cross-lingual encoders plus adapters. We find that these strategies perform especially well for classifying rare phenotypes and we advise on which method to prefer in which situation. Our results show that using multilingual data overall improves clinical phenotyping models and can compensate for data sparseness.
* LREC 2022 submmision: January 2022
What Do You See in this Patient? Behavioral Testing of Clinical NLP Models
Nov 30, 2021


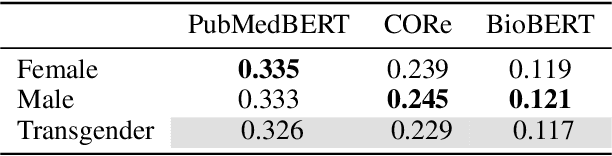
Abstract:Decision support systems based on clinical notes have the potential to improve patient care by pointing doctors towards overseen risks. Predicting a patient's outcome is an essential part of such systems, for which the use of deep neural networks has shown promising results. However, the patterns learned by these networks are mostly opaque and previous work revealed flaws regarding the reproduction of unintended biases. We thus introduce an extendable testing framework that evaluates the behavior of clinical outcome models regarding changes of the input. The framework helps to understand learned patterns and their influence on model decisions. In this work, we apply it to analyse the change in behavior with regard to the patient characteristics gender, age and ethnicity. Our evaluation of three current clinical NLP models demonstrates the concrete effects of these characteristics on the models' decisions. They show that model behavior varies drastically even when fine-tuned on the same data and that allegedly best-performing models have not always learned the most medically plausible patterns.
Self-supervised Answer Retrieval on Clinical Notes
Aug 02, 2021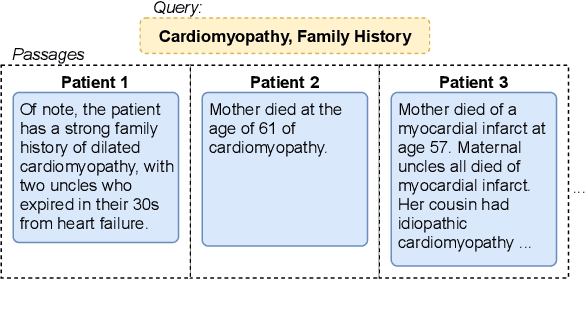
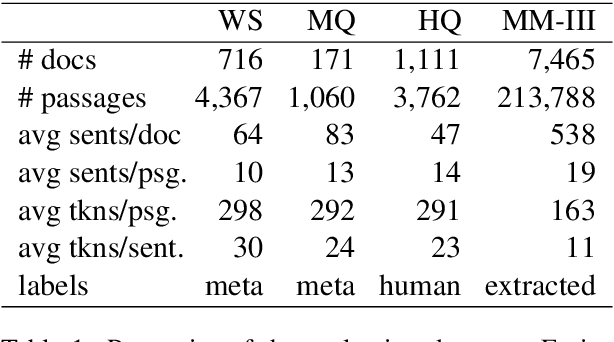
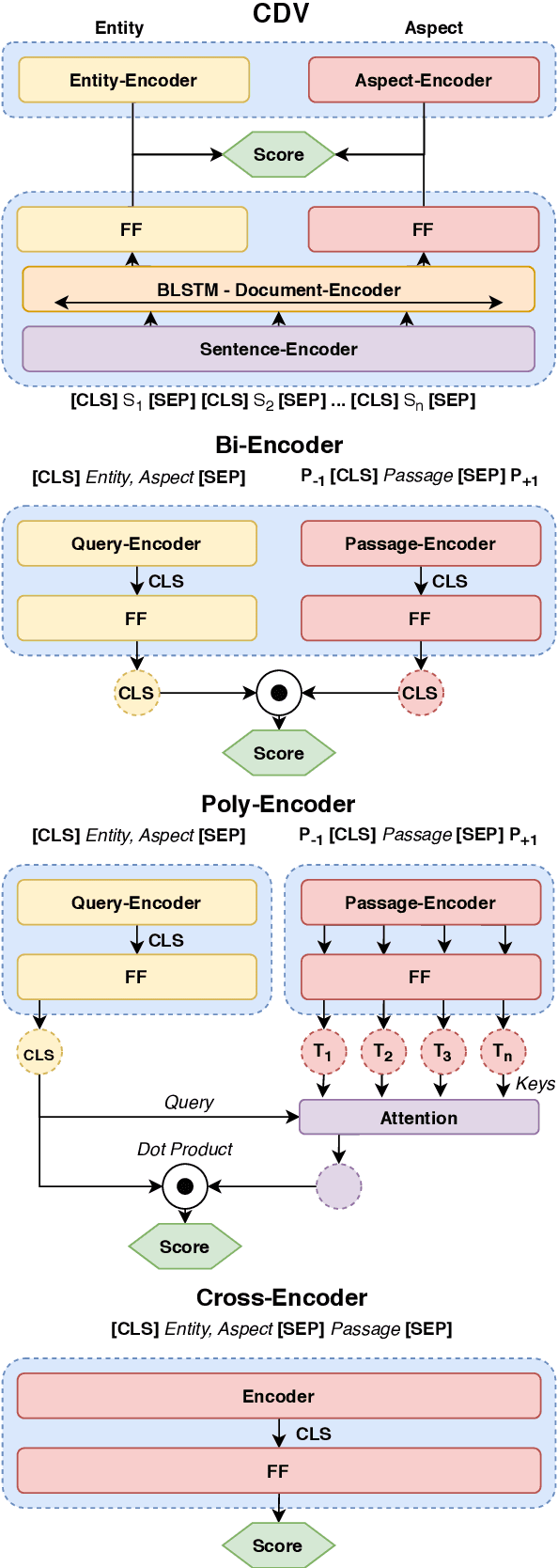
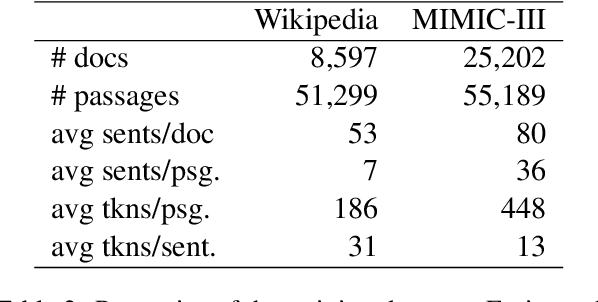
Abstract:Retrieving answer passages from long documents is a complex task requiring semantic understanding of both discourse and document context. We approach this challenge specifically in a clinical scenario, where doctors retrieve cohorts of patients based on diagnoses and other latent medical aspects. We introduce CAPR, a rule-based self-supervision objective for training Transformer language models for domain-specific passage matching. In addition, we contribute a novel retrieval dataset based on clinical notes to simulate this scenario on a large corpus of clinical notes. We apply our objective in four Transformer-based architectures: Contextual Document Vectors, Bi-, Poly- and Cross-encoders. From our extensive evaluation on MIMIC-III and three other healthcare datasets, we report that CAPR outperforms strong baselines in the retrieval of domain-specific passages and effectively generalizes across rule-based and human-labeled passages. This makes the model powerful especially in zero-shot scenarios where only limited training data is available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge