Paul Grundmann
MedAlpaca -- An Open-Source Collection of Medical Conversational AI Models and Training Data
Apr 14, 2023Abstract:As large language models (LLMs) like OpenAI's GPT series continue to make strides, we witness the emergence of artificial intelligence applications in an ever-expanding range of fields. In medicine, these LLMs hold considerable promise for improving medical workflows, diagnostics, patient care, and education. Yet, there is an urgent need for open-source models that can be deployed on-premises to safeguard patient privacy. In our work, we present an innovative dataset consisting of over 160,000 entries, specifically crafted to fine-tune LLMs for effective medical applications. We investigate the impact of fine-tuning these datasets on publicly accessible pre-trained LLMs, and subsequently, we juxtapose the performance of pre-trained-only models against the fine-tuned models concerning the examinations that future medical doctors must pass to achieve certification.
MEDBERT.de: A Comprehensive German BERT Model for the Medical Domain
Mar 24, 2023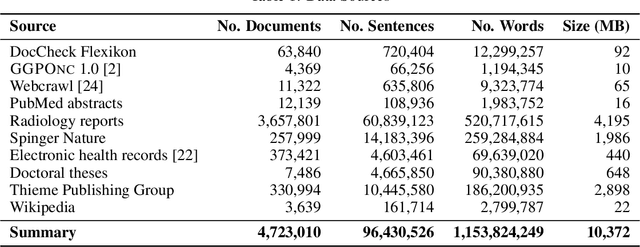
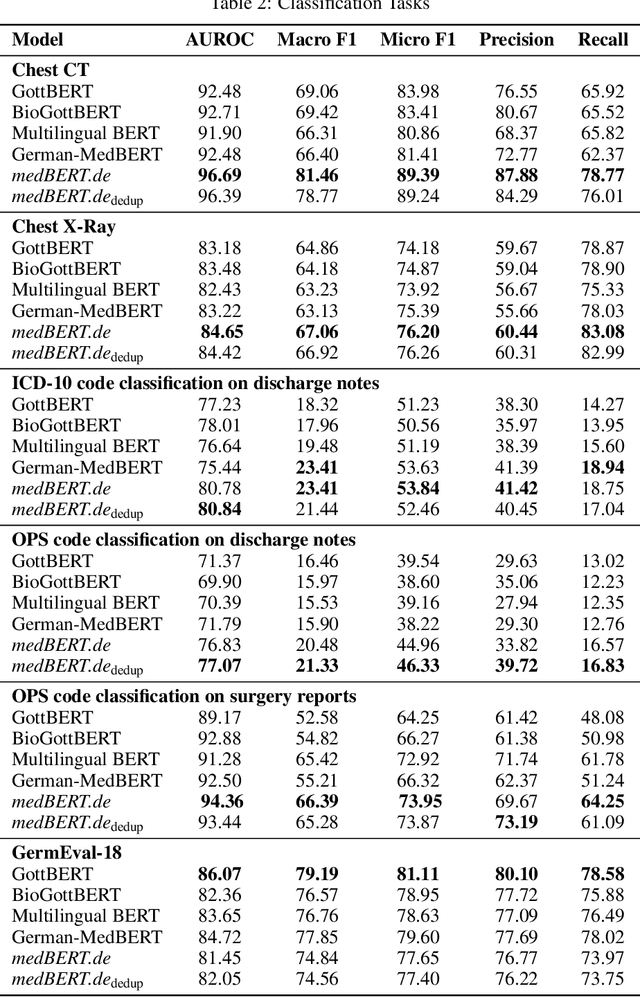
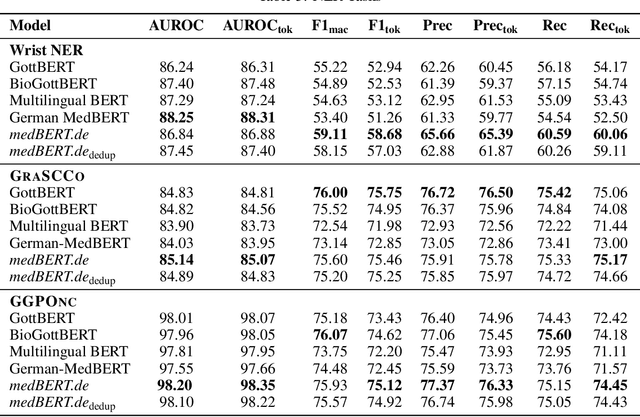
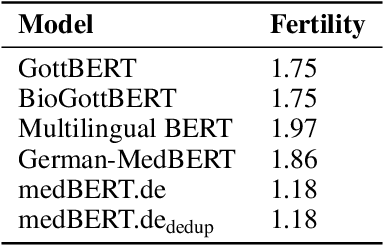
Abstract:This paper presents medBERTde, a pre-trained German BERT model specifically designed for the German medical domain. The model has been trained on a large corpus of 4.7 Million German medical documents and has been shown to achieve new state-of-the-art performance on eight different medical benchmarks covering a wide range of disciplines and medical document types. In addition to evaluating the overall performance of the model, this paper also conducts a more in-depth analysis of its capabilities. We investigate the impact of data deduplication on the model's performance, as well as the potential benefits of using more efficient tokenization methods. Our results indicate that domain-specific models such as medBERTde are particularly useful for longer texts, and that deduplication of training data does not necessarily lead to improved performance. Furthermore, we found that efficient tokenization plays only a minor role in improving model performance, and attribute most of the improved performance to the large amount of training data. To encourage further research, the pre-trained model weights and new benchmarks based on radiological data are made publicly available for use by the scientific community.
Cross-Lingual Knowledge Transfer for Clinical Phenotyping
Aug 03, 2022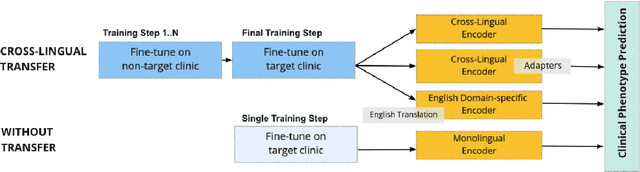

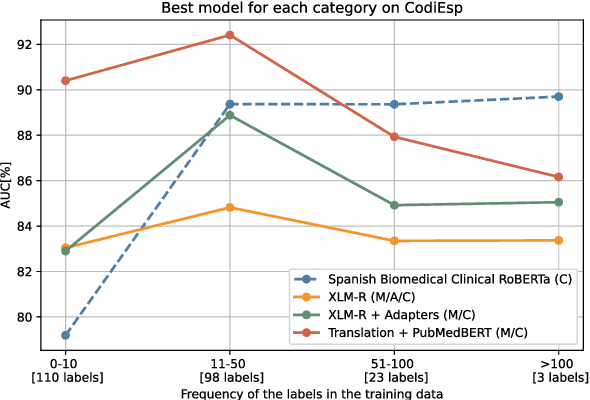
Abstract:Clinical phenotyping enables the automatic extraction of clinical conditions from patient records, which can be beneficial to doctors and clinics worldwide. However, current state-of-the-art models are mostly applicable to clinical notes written in English. We therefore investigate cross-lingual knowledge transfer strategies to execute this task for clinics that do not use the English language and have a small amount of in-domain data available. We evaluate these strategies for a Greek and a Spanish clinic leveraging clinical notes from different clinical domains such as cardiology, oncology and the ICU. Our results reveal two strategies that outperform the state-of-the-art: Translation-based methods in combination with domain-specific encoders and cross-lingual encoders plus adapters. We find that these strategies perform especially well for classifying rare phenotypes and we advise on which method to prefer in which situation. Our results show that using multilingual data overall improves clinical phenotyping models and can compensate for data sparseness.
* LREC 2022 submmision: January 2022
Self-supervised Answer Retrieval on Clinical Notes
Aug 02, 2021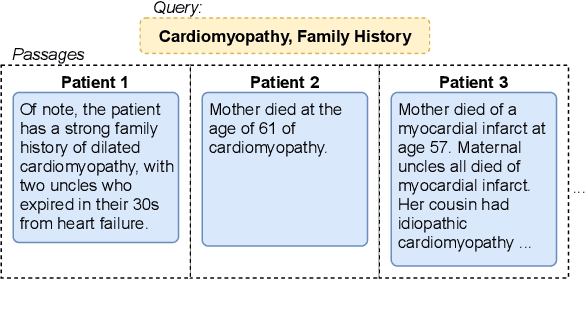
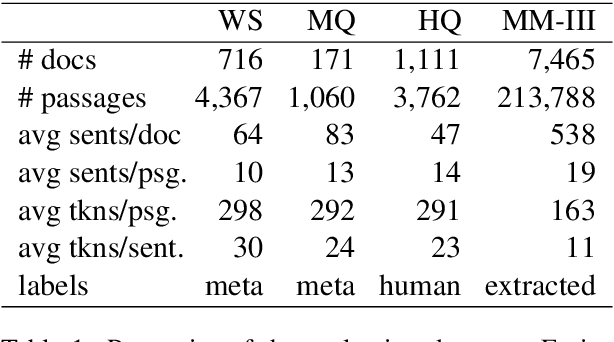
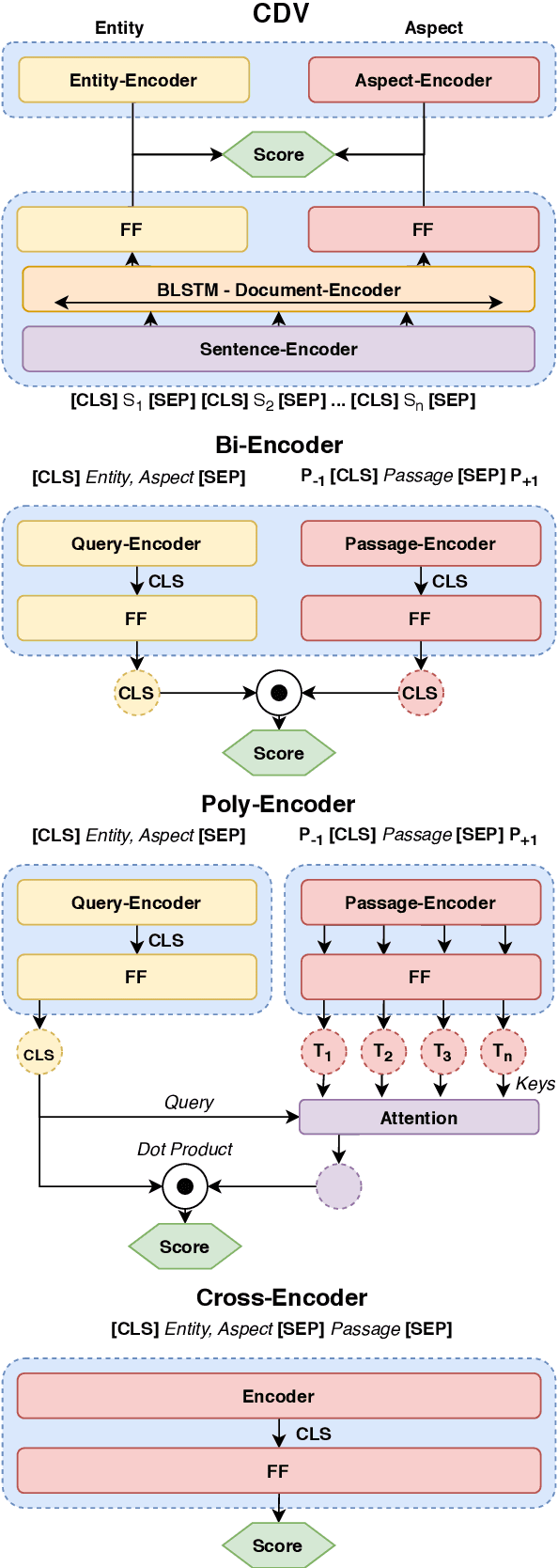
Abstract:Retrieving answer passages from long documents is a complex task requiring semantic understanding of both discourse and document context. We approach this challenge specifically in a clinical scenario, where doctors retrieve cohorts of patients based on diagnoses and other latent medical aspects. We introduce CAPR, a rule-based self-supervision objective for training Transformer language models for domain-specific passage matching. In addition, we contribute a novel retrieval dataset based on clinical notes to simulate this scenario on a large corpus of clinical notes. We apply our objective in four Transformer-based architectures: Contextual Document Vectors, Bi-, Poly- and Cross-encoders. From our extensive evaluation on MIMIC-III and three other healthcare datasets, we report that CAPR outperforms strong baselines in the retrieval of domain-specific passages and effectively generalizes across rule-based and human-labeled passages. This makes the model powerful especially in zero-shot scenarios where only limited training data is available.
Learning Contextualized Document Representations for Healthcare Answer Retrieval
Feb 03, 2020
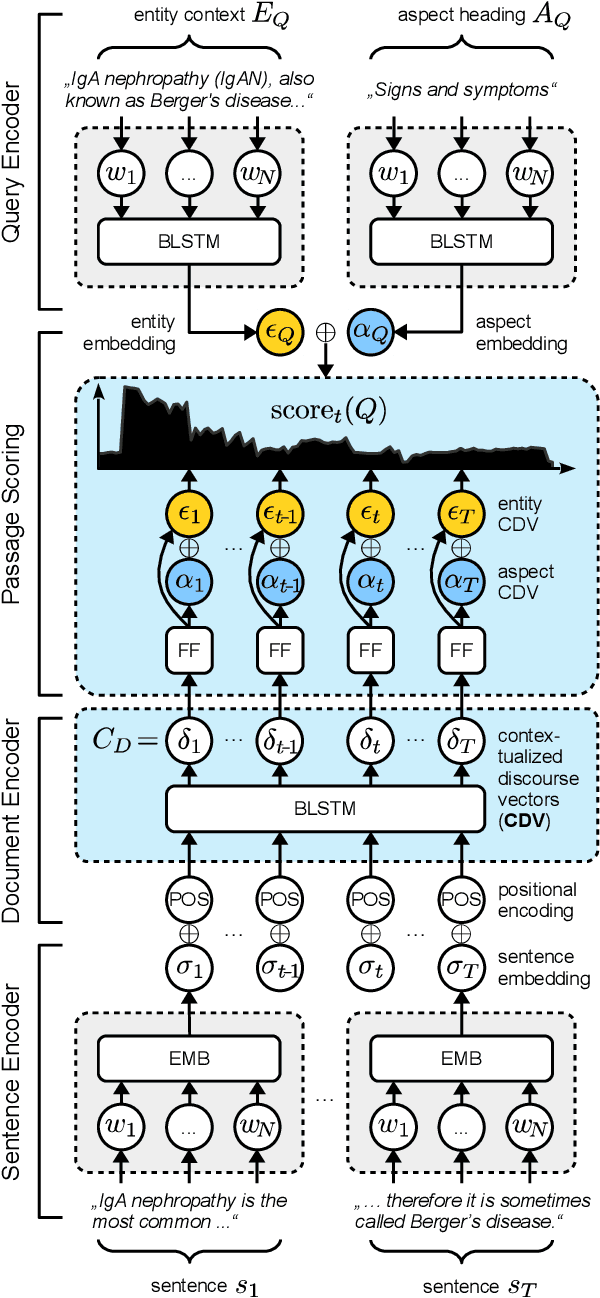
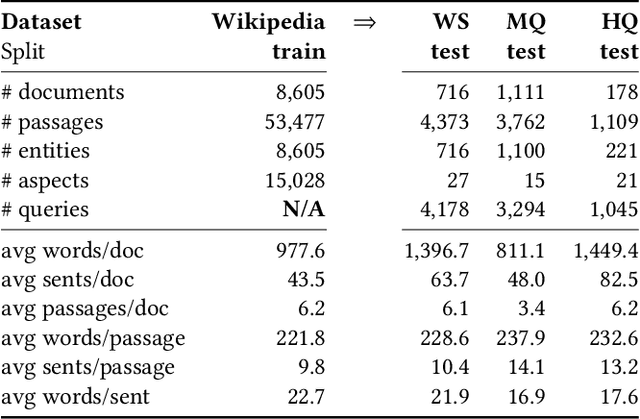
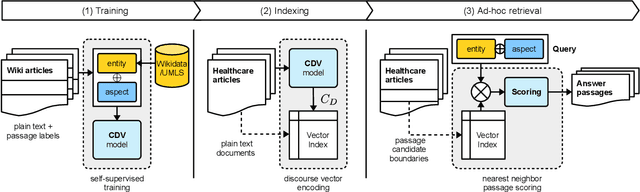
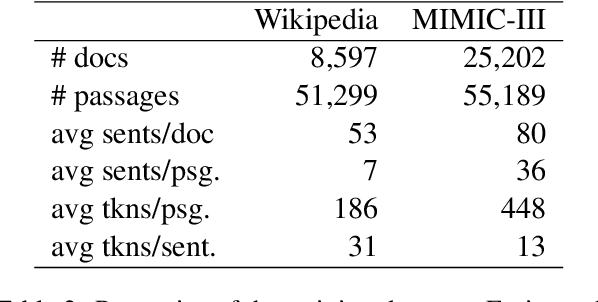
Abstract:We present Contextual Discourse Vectors (CDV), a distributed document representation for efficient answer retrieval from long healthcare documents. Our approach is based on structured query tuples of entities and aspects from free text and medical taxonomies. Our model leverages a dual encoder architecture with hierarchical LSTM layers and multi-task training to encode the position of clinical entities and aspects alongside the document discourse. We use our continuous representations to resolve queries with short latency using approximate nearest neighbor search on sentence level. We apply the CDV model for retrieving coherent answer passages from nine English public health resources from the Web, addressing both patients and medical professionals. Because there is no end-to-end training data available for all application scenarios, we train our model with self-supervised data from Wikipedia. We show that our generalized model significantly outperforms several state-of-the-art baselines for healthcare passage ranking and is able to adapt to heterogeneous domains without additional fine-tuning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge