Jens Kleesiek
Less Finetuning, Better Retrieval: Rethinking LLM Adaptation for Biomedical Retrievers via Synthetic Data and Model Merging
Feb 04, 2026Abstract:Retrieval-augmented generation (RAG) has become the backbone of grounding Large Language Models (LLMs), improving knowledge updates and reducing hallucinations. Recently, LLM-based retriever models have shown state-of-the-art performance for RAG applications. However, several technical aspects remain underexplored on how to adapt general-purpose LLMs into effective domain-specific retrievers, especially in specialized domains such as biomedicine. We present Synthesize-Train-Merge (STM), a modular framework that enhances decoder-only LLMs with synthetic hard negatives, retrieval prompt optimization, and model merging. Experiments on a subset of 12 medical and general tasks from the MTEB benchmark show STM boosts task-specific experts by up to 23.5\% (average 7.5\%) and produces merged models that outperform both single experts and strong baselines without extensive pretraining. Our results demonstrate a scalable, efficient path for turning general LLMs into high-performing, domain-specialized retrievers, preserving general-domain capabilities while excelling on specialized tasks.
AIANO: Enhancing Information Retrieval with AI-Augmented Annotation
Feb 04, 2026Abstract:The rise of Large Language Models (LLMs) and Retrieval-Augmented Generation (RAG) has rapidly increased the need for high-quality, curated information retrieval datasets. These datasets, however, are currently created with off-the-shelf annotation tools that make the annotation process complex and inefficient. To streamline this process, we developed a specialized annotation tool - AIANO. By adopting an AI-augmented annotation workflow that tightly integrates human expertise with LLM assistance, AIANO enables annotators to leverage AI suggestions while retaining full control over annotation decisions. In a within-subject user study ($n = 15$), participants created question-answering datasets using both a baseline tool and AIANO. AIANO nearly doubled annotation speed compared to the baseline while being easier to use and improving retrieval accuracy. These results demonstrate that AIANO's AI-augmented approach accelerates and enhances dataset creation for information retrieval tasks, advancing annotation capabilities in retrieval-intensive domains.
Region-Normalized DPO for Medical Image Segmentation under Noisy Judges
Jan 30, 2026Abstract:While dense pixel-wise annotations remain the gold standard for medical image segmentation, they are costly to obtain and limit scalability. In contrast, many deployed systems already produce inexpensive automatic quality-control (QC) signals like model agreement, uncertainty measures, or learned mask-quality scores which can be used for further model training without additional ground-truth annotation. However, these signals can be noisy and biased, making preference-based fine-tuning susceptible to harmful updates. We study Direct Preference Optimization (DPO) for segmentation from such noisy judges using proposals generated by a supervised base segmenter trained on a small labeled set. We find that outcomes depend strongly on how preference pairs are mined: selecting the judge's top-ranked proposal can improve peak performance when the judge is reliable, but can amplify harmful errors under weaker judges. We propose Region-Normalized DPO (RN-DPO), a segmentation-aware objective which normalizes preference updates by the size of the disagreement region between masks, reducing the leverage of harmful comparisons and improving optimization stability. Across two medical datasets and multiple regimes, RN-DPO improves sustained performance and stabilizes preference-based fine-tuning, outperforming standard DPO and strong baselines without requiring additional pixel annotations.
Efficient Complex-Valued Vision Transformers for MRI Classification Directly from k-Space
Jan 26, 2026Abstract:Deep learning applications in Magnetic Resonance Imaging (MRI) predominantly operate on reconstructed magnitude images, a process that discards phase information and requires computationally expensive transforms. Standard neural network architectures rely on local operations (convolutions or grid-patches) that are ill-suited for the global, non-local nature of raw frequency-domain (k-Space) data. In this work, we propose a novel complex-valued Vision Transformer (kViT) designed to perform classification directly on k-Space data. To bridge the geometric disconnect between current architectures and MRI physics, we introduce a radial k-Space patching strategy that respects the spectral energy distribution of the frequency-domain. Extensive experiments on the fastMRI and in-house datasets demonstrate that our approach achieves classification performance competitive with state-of-the-art image-domain baselines (ResNet, EfficientNet, ViT). Crucially, kViT exhibits superior robustness to high acceleration factors and offers a paradigm shift in computational efficiency, reducing VRAM consumption during training by up to 68$\times$ compared to standard methods. This establishes a pathway for resource-efficient, direct-from-scanner AI analysis.
CT-GRAPH: Hierarchical Graph Attention Network for Anatomy-Guided CT Report Generation
Aug 07, 2025Abstract:As medical imaging is central to diagnostic processes, automating the generation of radiology reports has become increasingly relevant to assist radiologists with their heavy workloads. Most current methods rely solely on global image features, failing to capture fine-grained organ relationships crucial for accurate reporting. To this end, we propose CT-GRAPH, a hierarchical graph attention network that explicitly models radiological knowledge by structuring anatomical regions into a graph, linking fine-grained organ features to coarser anatomical systems and a global patient context. Our method leverages pretrained 3D medical feature encoders to obtain global and organ-level features by utilizing anatomical masks. These features are further refined within the graph and then integrated into a large language model to generate detailed medical reports. We evaluate our approach for the task of report generation on the large-scale chest CT dataset CT-RATE. We provide an in-depth analysis of pretrained feature encoders for CT report generation and show that our method achieves a substantial improvement of absolute 7.9\% in F1 score over current state-of-the-art methods. The code is publicly available at https://github.com/hakal104/CT-GRAPH.
Automatic Fine-grained Segmentation-assisted Report Generation
Jul 22, 2025Abstract:Reliable end-to-end clinical report generation has been a longstanding goal of medical ML research. The end goal for this process is to alleviate radiologists' workloads and provide second opinions to clinicians or patients. Thus, a necessary prerequisite for report generation models is a strong general performance and some type of innate grounding capability, to convince clinicians or patients of the veracity of the generated reports. In this paper, we present ASaRG (\textbf{A}utomatic \textbf{S}egmentation-\textbf{a}ssisted \textbf{R}eport \textbf{G}eneration), an extension of the popular LLaVA architecture that aims to tackle both of these problems. ASaRG proposes to fuse intermediate features and fine-grained segmentation maps created by specialist radiological models into LLaVA's multi-modal projection layer via simple concatenation. With a small number of added parameters, our approach achieves a +0.89\% performance gain ($p=0.012$) in CE F1 score compared to the LLaVA baseline when using only intermediate features, and +2.77\% performance gain ($p<0.001$) when adding a combination of intermediate features and fine-grained segmentation maps. Compared with COMG and ORID, two other report generation methods that utilize segmentations, the performance gain amounts to 6.98\% and 6.28\% in F1 score, respectively. ASaRG is not mutually exclusive with other changes made to the LLaVA architecture, potentially allowing our method to be combined with other advances in the field. Finally, the use of an arbitrary number of segmentations as part of the input demonstrably allows tracing elements of the report to the corresponding segmentation maps and verifying the groundedness of assessments. Our code will be made publicly available at a later date.
Enhancing Privacy: The Utility of Stand-Alone Synthetic CT and MRI for Tumor and Bone Segmentation
Jun 13, 2025Abstract:AI requires extensive datasets, while medical data is subject to high data protection. Anonymization is essential, but poses a challenge for some regions, such as the head, as identifying structures overlap with regions of clinical interest. Synthetic data offers a potential solution, but studies often lack rigorous evaluation of realism and utility. Therefore, we investigate to what extent synthetic data can replace real data in segmentation tasks. We employed head and neck cancer CT scans and brain glioma MRI scans from two large datasets. Synthetic data were generated using generative adversarial networks and diffusion models. We evaluated the quality of the synthetic data using MAE, MS-SSIM, Radiomics and a Visual Turing Test (VTT) performed by 5 radiologists and their usefulness in segmentation tasks using DSC. Radiomics indicates high fidelity of synthetic MRIs, but fall short in producing highly realistic CT tissue, with correlation coefficient of 0.8784 and 0.5461 for MRI and CT tumors, respectively. DSC results indicate limited utility of synthetic data: tumor segmentation achieved DSC=0.064 on CT and 0.834 on MRI, while bone segmentation a mean DSC=0.841. Relation between DSC and correlation is observed, but is limited by the complexity of the task. VTT results show synthetic CTs' utility, but with limited educational applications. Synthetic data can be used independently for the segmentation task, although limited by the complexity of the structures to segment. Advancing generative models to better tolerate heterogeneous inputs and learn subtle details is essential for enhancing their realism and expanding their application potential.
Good Enough: Is it Worth Improving your Label Quality?
May 27, 2025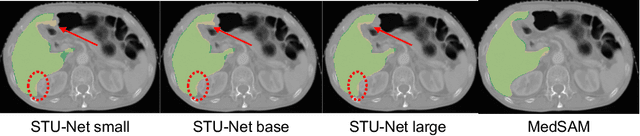
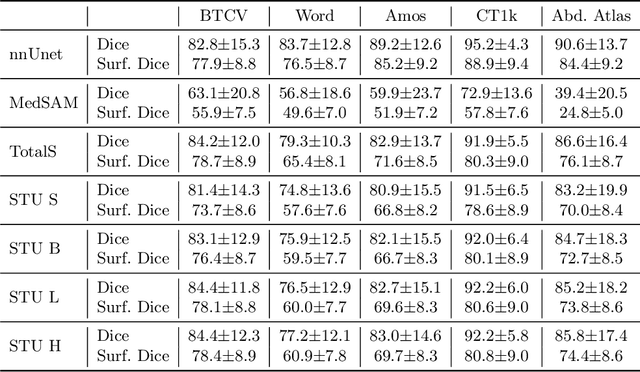
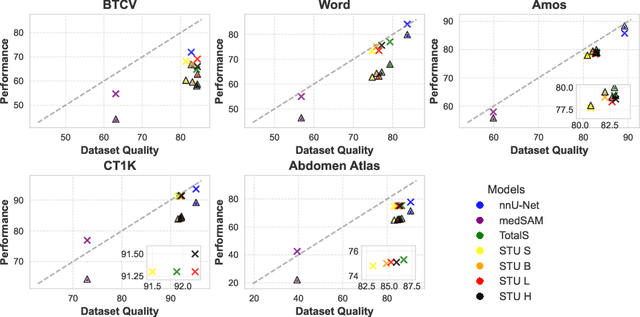
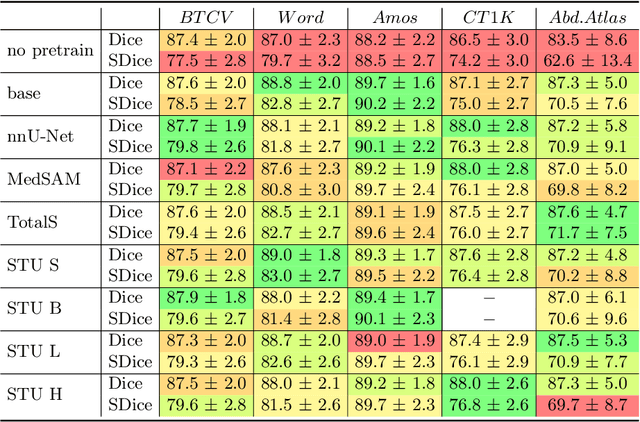
Abstract:Improving label quality in medical image segmentation is costly, but its benefits remain unclear. We systematically evaluate its impact using multiple pseudo-labeled versions of CT datasets, generated by models like nnU-Net, TotalSegmentator, and MedSAM. Our results show that while higher-quality labels improve in-domain performance, gains remain unclear if below a small threshold. For pre-training, label quality has minimal impact, suggesting that models rather transfer general concepts than detailed annotations. These findings provide guidance on when improving label quality is worth the effort.
A Modular Approach for Clinical SLMs Driven by Synthetic Data with Pre-Instruction Tuning, Model Merging, and Clinical-Tasks Alignment
May 15, 2025Abstract:High computation costs and latency of large language models such as GPT-4 have limited their deployment in clinical settings. Small language models (SLMs) offer a cost-effective alternative, but their limited capacity requires biomedical domain adaptation, which remains challenging. An additional bottleneck is the unavailability and high sensitivity of clinical data. To address these challenges, we propose a novel framework for adapting SLMs into high-performing clinical models. We introduce the MediPhi collection of 3.8B-parameter SLMs developed with our novel framework: pre-instruction tuning of experts on relevant medical and clinical corpora (PMC, Medical Guideline, MedWiki, etc.), model merging, and clinical-tasks alignment. To cover most clinical tasks, we extended the CLUE benchmark to CLUE+, doubling its size. Our expert models deliver relative improvements on this benchmark over the base model without any task-specific fine-tuning: 64.3% on medical entities, 49.5% on radiology reports, and 44% on ICD-10 coding (outperforming GPT-4-0125 by 14%). We unify the expert models into MediPhi via model merging, preserving gains across benchmarks. Furthermore, we built the MediFlow collection, a synthetic dataset of 2.5 million high-quality instructions on 14 medical NLP tasks, 98 fine-grained document types, and JSON format support. Alignment of MediPhi using supervised fine-tuning and direct preference optimization achieves further gains of 18.9% on average.
PhaseGen: A Diffusion-Based Approach for Complex-Valued MRI Data Generation
Apr 10, 2025Abstract:Magnetic resonance imaging (MRI) raw data, or k-Space data, is complex-valued, containing both magnitude and phase information. However, clinical and existing Artificial Intelligence (AI)-based methods focus only on magnitude images, discarding the phase data despite its potential for downstream tasks, such as tumor segmentation and classification. In this work, we introduce $\textit{PhaseGen}$, a novel complex-valued diffusion model for generating synthetic MRI raw data conditioned on magnitude images, commonly used in clinical practice. This enables the creation of artificial complex-valued raw data, allowing pretraining for models that require k-Space information. We evaluate PhaseGen on two tasks: skull-stripping directly in k-Space and MRI reconstruction using the publicly available FastMRI dataset. Our results show that training with synthetic phase data significantly improves generalization for skull-stripping on real-world data, with an increased segmentation accuracy from $41.1\%$ to $80.1\%$, and enhances MRI reconstruction when combined with limited real-world data. This work presents a step forward in utilizing generative AI to bridge the gap between magnitude-based datasets and the complex-valued nature of MRI raw data. This approach allows researchers to leverage the vast amount of avaliable image domain data in combination with the information-rich k-Space data for more accurate and efficient diagnostic tasks. We make our code publicly $\href{https://github.com/TIO-IKIM/PhaseGen}{\text{available here}}$.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge