Julius Keyl
PathMR: Multimodal Visual Reasoning for Interpretable Pathology Diagnosis
Aug 28, 2025Abstract:Deep learning based automated pathological diagnosis has markedly improved diagnostic efficiency and reduced variability between observers, yet its clinical adoption remains limited by opaque model decisions and a lack of traceable rationale. To address this, recent multimodal visual reasoning architectures provide a unified framework that generates segmentation masks at the pixel level alongside semantically aligned textual explanations. By localizing lesion regions and producing expert style diagnostic narratives, these models deliver the transparent and interpretable insights necessary for dependable AI assisted pathology. Building on these advancements, we propose PathMR, a cell-level Multimodal visual Reasoning framework for Pathological image analysis. Given a pathological image and a textual query, PathMR generates expert-level diagnostic explanations while simultaneously predicting cell distribution patterns. To benchmark its performance, we evaluated our approach on the publicly available PathGen dataset as well as on our newly developed GADVR dataset. Extensive experiments on these two datasets demonstrate that PathMR consistently outperforms state-of-the-art visual reasoning methods in text generation quality, segmentation accuracy, and cross-modal alignment. These results highlight the potential of PathMR for improving interpretability in AI-driven pathological diagnosis. The code will be publicly available in https://github.com/zhangye-zoe/PathMR.
CellViT++: Energy-Efficient and Adaptive Cell Segmentation and Classification Using Foundation Models
Jan 09, 2025



Abstract:Digital Pathology is a cornerstone in the diagnosis and treatment of diseases. A key task in this field is the identification and segmentation of cells in hematoxylin and eosin-stained images. Existing methods for cell segmentation often require extensive annotated datasets for training and are limited to a predefined cell classification scheme. To overcome these limitations, we propose $\text{CellViT}^{{\scriptscriptstyle ++}}$, a framework for generalized cell segmentation in digital pathology. $\text{CellViT}^{{\scriptscriptstyle ++}}$ utilizes Vision Transformers with foundation models as encoders to compute deep cell features and segmentation masks simultaneously. To adapt to unseen cell types, we rely on a computationally efficient approach. It requires minimal data for training and leads to a drastically reduced carbon footprint. We demonstrate excellent performance on seven different datasets, covering a broad spectrum of cell types, organs, and clinical settings. The framework achieves remarkable zero-shot segmentation and data-efficient cell-type classification. Furthermore, we show that $\text{CellViT}^{{\scriptscriptstyle ++}}$ can leverage immunofluorescence stainings to generate training datasets without the need for pathologist annotations. The automated dataset generation approach surpasses the performance of networks trained on manually labeled data, demonstrating its effectiveness in creating high-quality training datasets without expert annotations. To advance digital pathology, $\text{CellViT}^{{\scriptscriptstyle ++}}$ is available as an open-source framework featuring a user-friendly, web-based interface for visualization and annotation. The code is available under https://github.com/TIO-IKIM/CellViT-plus-plus.
MedShapeNet -- A Large-Scale Dataset of 3D Medical Shapes for Computer Vision
Sep 12, 2023



Abstract:We present MedShapeNet, a large collection of anatomical shapes (e.g., bones, organs, vessels) and 3D surgical instrument models. Prior to the deep learning era, the broad application of statistical shape models (SSMs) in medical image analysis is evidence that shapes have been commonly used to describe medical data. Nowadays, however, state-of-the-art (SOTA) deep learning algorithms in medical imaging are predominantly voxel-based. In computer vision, on the contrary, shapes (including, voxel occupancy grids, meshes, point clouds and implicit surface models) are preferred data representations in 3D, as seen from the numerous shape-related publications in premier vision conferences, such as the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), as well as the increasing popularity of ShapeNet (about 51,300 models) and Princeton ModelNet (127,915 models) in computer vision research. MedShapeNet is created as an alternative to these commonly used shape benchmarks to facilitate the translation of data-driven vision algorithms to medical applications, and it extends the opportunities to adapt SOTA vision algorithms to solve critical medical problems. Besides, the majority of the medical shapes in MedShapeNet are modeled directly on the imaging data of real patients, and therefore it complements well existing shape benchmarks comprising of computer-aided design (CAD) models. MedShapeNet currently includes more than 100,000 medical shapes, and provides annotations in the form of paired data. It is therefore also a freely available repository of 3D models for extended reality (virtual reality - VR, augmented reality - AR, mixed reality - MR) and medical 3D printing. This white paper describes in detail the motivations behind MedShapeNet, the shape acquisition procedures, the use cases, as well as the usage of the online shape search portal: https://medshapenet.ikim.nrw/
CellViT: Vision Transformers for Precise Cell Segmentation and Classification
Jun 27, 2023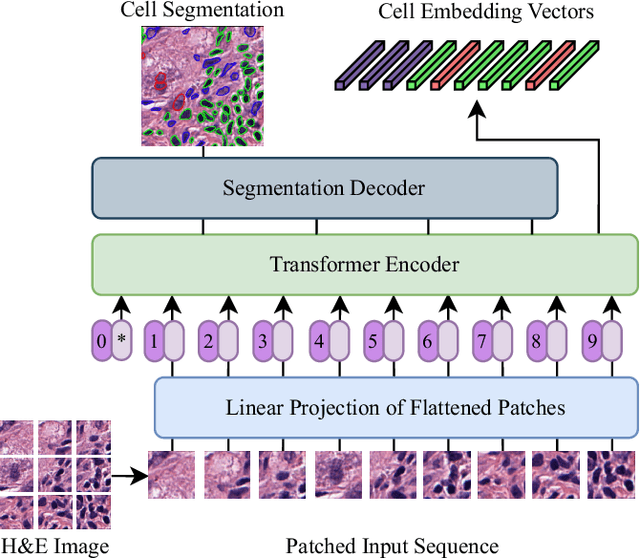
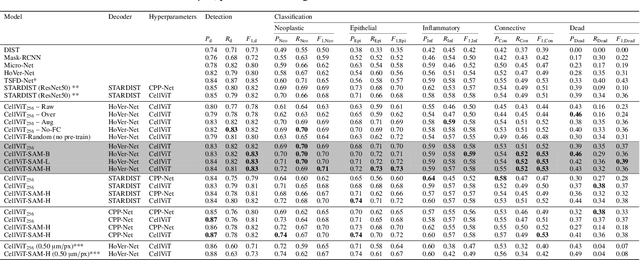
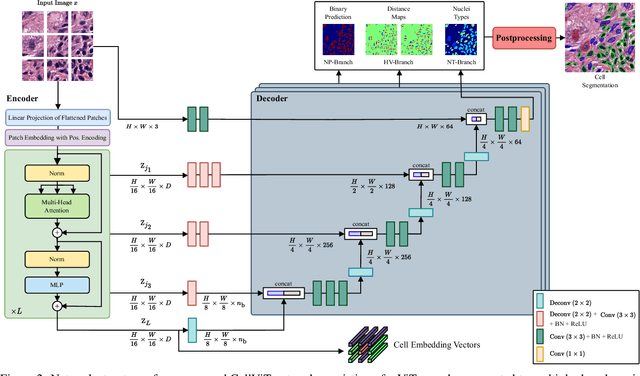

Abstract:Nuclei detection and segmentation in hematoxylin and eosin-stained (H&E) tissue images are important clinical tasks and crucial for a wide range of applications. However, it is a challenging task due to nuclei variances in staining and size, overlapping boundaries, and nuclei clustering. While convolutional neural networks have been extensively used for this task, we explore the potential of Transformer-based networks in this domain. Therefore, we introduce a new method for automated instance segmentation of cell nuclei in digitized tissue samples using a deep learning architecture based on Vision Transformer called CellViT. CellViT is trained and evaluated on the PanNuke dataset, which is one of the most challenging nuclei instance segmentation datasets, consisting of nearly 200,000 annotated Nuclei into 5 clinically important classes in 19 tissue types. We demonstrate the superiority of large-scale in-domain and out-of-domain pre-trained Vision Transformers by leveraging the recently published Segment Anything Model and a ViT-encoder pre-trained on 104 million histological image patches - achieving state-of-the-art nuclei detection and instance segmentation performance on the PanNuke dataset with a mean panoptic quality of 0.51 and an F1-detection score of 0.83. The code is publicly available at https://github.com/TIO-IKIM/CellViT
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge