Gijs Luijten
Efficient MedSAMs: Segment Anything in Medical Images on Laptop
Dec 20, 2024


Abstract:Promptable segmentation foundation models have emerged as a transformative approach to addressing the diverse needs in medical images, but most existing models require expensive computing, posing a big barrier to their adoption in clinical practice. In this work, we organized the first international competition dedicated to promptable medical image segmentation, featuring a large-scale dataset spanning nine common imaging modalities from over 20 different institutions. The top teams developed lightweight segmentation foundation models and implemented an efficient inference pipeline that substantially reduced computational requirements while maintaining state-of-the-art segmentation accuracy. Moreover, the post-challenge phase advanced the algorithms through the design of performance booster and reproducibility tasks, resulting in improved algorithms and validated reproducibility of the winning solution. Furthermore, the best-performing algorithms have been incorporated into the open-source software with a user-friendly interface to facilitate clinical adoption. The data and code are publicly available to foster the further development of medical image segmentation foundation models and pave the way for impactful real-world applications.
Comparative Analysis of nnUNet and MedNeXt for Head and Neck Tumor Segmentation in MRI-guided Radiotherapy
Nov 22, 2024Abstract:Radiation therapy (RT) is essential in treating head and neck cancer (HNC), with magnetic resonance imaging(MRI)-guided RT offering superior soft tissue contrast and functional imaging. However, manual tumor segmentation is time-consuming and complex, and therfore remains a challenge. In this study, we present our solution as team TUMOR to the HNTS-MRG24 MICCAI Challenge which is focused on automated segmentation of primary gross tumor volumes (GTVp) and metastatic lymph node gross tumor volume (GTVn) in pre-RT and mid-RT MRI images. We utilized the HNTS-MRG2024 dataset, which consists of 150 MRI scans from patients diagnosed with HNC, including original and registered pre-RT and mid-RT T2-weighted images with corresponding segmentation masks for GTVp and GTVn. We employed two state-of-the-art models in deep learning, nnUNet and MedNeXt. For Task 1, we pretrained models on pre-RT registered and mid-RT images, followed by fine-tuning on original pre-RT images. For Task 2, we combined registered pre-RT images, registered pre-RT segmentation masks, and mid-RT data as a multi-channel input for training. Our solution for Task 1 achieved 1st place in the final test phase with an aggregated Dice Similarity Coefficient of 0.8254, and our solution for Task 2 ranked 8th with a score of 0.7005. The proposed solution is publicly available at Github Repository.
Brain Tumour Removing and Missing Modality Generation using 3D WDM
Nov 07, 2024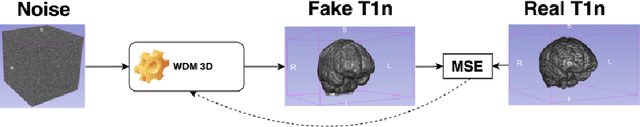
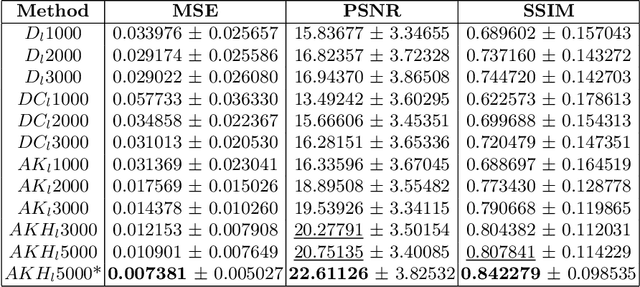
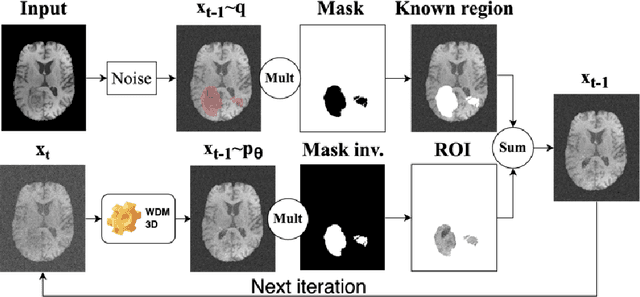
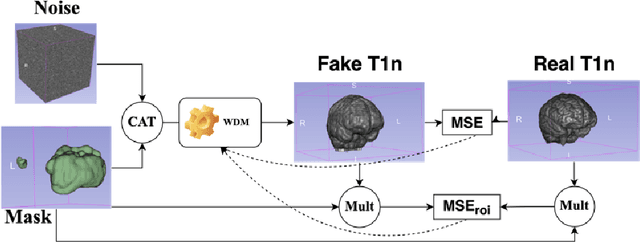
Abstract:This paper presents the second-placed solution for task 8 and the participation solution for task 7 of BraTS 2024. The adoption of automated brain analysis algorithms to support clinical practice is increasing. However, many of these algorithms struggle with the presence of brain lesions or the absence of certain MRI modalities. The alterations in the brain's morphology leads to high variability and thus poor performance of predictive models that were trained only on healthy brains. The lack of information that is usually provided by some of the missing MRI modalities also reduces the reliability of the prediction models trained with all modalities. In order to improve the performance of these models, we propose the use of conditional 3D wavelet diffusion models. The wavelet transform enabled full-resolution image training and prediction on a GPU with 48 GB VRAM, without patching or downsampling, preserving all information for prediction. For the inpainting task of BraTS 2024, the use of a large and variable number of healthy masks and the stability and efficiency of the 3D wavelet diffusion model resulted in 0.007, 22.61 and 0.842 in the validation set and 0.07 , 22.8 and 0.91 in the testing set (MSE, PSNR and SSIM respectively). The code for these tasks is available at https://github.com/ShadowTwin41/BraTS_2023_2024_solutions.
Spacewalker: Traversing Representation Spaces for Fast Interactive Exploration and Annotation of Unstructured Data
Sep 25, 2024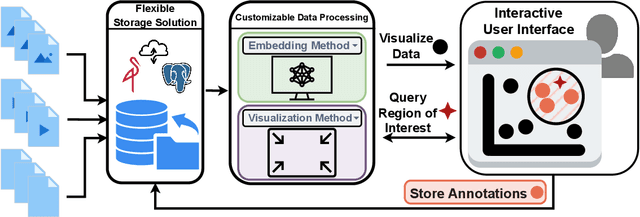
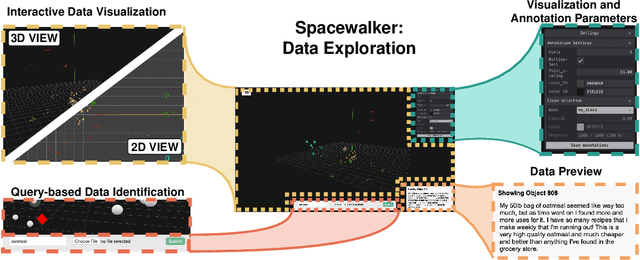
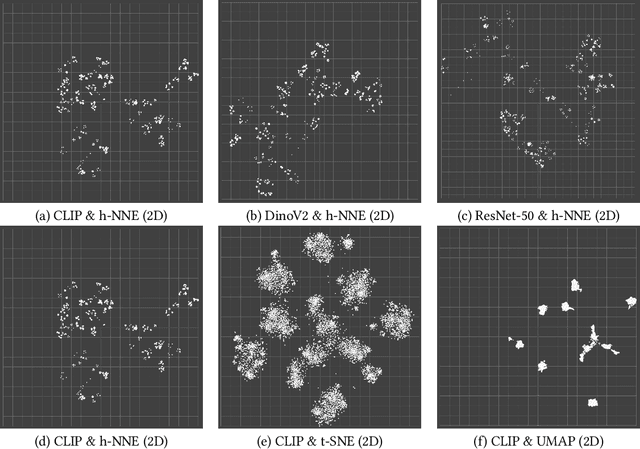
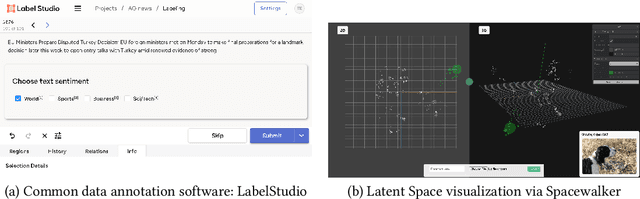
Abstract:Unstructured data in industries such as healthcare, finance, and manufacturing presents significant challenges for efficient analysis and decision making. Detecting patterns within this data and understanding their impact is critical but complex without the right tools. Traditionally, these tasks relied on the expertise of data analysts or labor-intensive manual reviews. In response, we introduce Spacewalker, an interactive tool designed to explore and annotate data across multiple modalities. Spacewalker allows users to extract data representations and visualize them in low-dimensional spaces, enabling the detection of semantic similarities. Through extensive user studies, we assess Spacewalker's effectiveness in data annotation and integrity verification. Results show that the tool's ability to traverse latent spaces and perform multi-modal queries significantly enhances the user's capacity to quickly identify relevant data. Moreover, Spacewalker allows for annotation speed-ups far superior to conventional methods, making it a promising tool for efficiently navigating unstructured data and improving decision making processes. The code of this work is open-source and can be found at: https://github.com/code-lukas/Spacewalker
Deep PCCT: Photon Counting Computed Tomography Deep Learning Applications Review
Feb 06, 2024
Abstract:Medical imaging faces challenges such as limited spatial resolution, interference from electronic noise and poor contrast-to-noise ratios. Photon Counting Computed Tomography (PCCT) has emerged as a solution, addressing these issues with its innovative technology. This review delves into the recent developments and applications of PCCT in pre-clinical research, emphasizing its potential to overcome traditional imaging limitations. For example PCCT has demonstrated remarkable efficacy in improving the detection of subtle abnormalities in breast, providing a level of detail previously unattainable. Examining the current literature on PCCT, it presents a comprehensive analysis of the technology, highlighting the main features of scanners and their varied applications. In addition, it explores the integration of deep learning into PCCT, along with the study of radiomic features, presenting successful applications in data processing. While acknowledging these advances, it also discusses the existing challenges in this field, paving the way for future research and improvements in medical imaging technologies. Despite the limited number of articles on this subject, due to the recent integration of PCCT at a clinical level, its potential benefits extend to various diagnostic applications.
Multilingual Natural Language Processing Model for Radiology Reports -- The Summary is all you need!
Oct 06, 2023



Abstract:The impression section of a radiology report summarizes important radiology findings and plays a critical role in communicating these findings to physicians. However, the preparation of these summaries is time-consuming and error-prone for radiologists. Recently, numerous models for radiology report summarization have been developed. Nevertheless, there is currently no model that can summarize these reports in multiple languages. Such a model could greatly improve future research and the development of Deep Learning models that incorporate data from patients with different ethnic backgrounds. In this study, the generation of radiology impressions in different languages was automated by fine-tuning a model, publicly available, based on a multilingual text-to-text Transformer to summarize findings available in English, Portuguese, and German radiology reports. In a blind test, two board-certified radiologists indicated that for at least 70% of the system-generated summaries, the quality matched or exceeded the corresponding human-written summaries, suggesting substantial clinical reliability. Furthermore, this study showed that the multilingual model outperformed other models that specialized in summarizing radiology reports in only one language, as well as models that were not specifically designed for summarizing radiology reports, such as ChatGPT.
MedShapeNet -- A Large-Scale Dataset of 3D Medical Shapes for Computer Vision
Sep 12, 2023



Abstract:We present MedShapeNet, a large collection of anatomical shapes (e.g., bones, organs, vessels) and 3D surgical instrument models. Prior to the deep learning era, the broad application of statistical shape models (SSMs) in medical image analysis is evidence that shapes have been commonly used to describe medical data. Nowadays, however, state-of-the-art (SOTA) deep learning algorithms in medical imaging are predominantly voxel-based. In computer vision, on the contrary, shapes (including, voxel occupancy grids, meshes, point clouds and implicit surface models) are preferred data representations in 3D, as seen from the numerous shape-related publications in premier vision conferences, such as the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), as well as the increasing popularity of ShapeNet (about 51,300 models) and Princeton ModelNet (127,915 models) in computer vision research. MedShapeNet is created as an alternative to these commonly used shape benchmarks to facilitate the translation of data-driven vision algorithms to medical applications, and it extends the opportunities to adapt SOTA vision algorithms to solve critical medical problems. Besides, the majority of the medical shapes in MedShapeNet are modeled directly on the imaging data of real patients, and therefore it complements well existing shape benchmarks comprising of computer-aided design (CAD) models. MedShapeNet currently includes more than 100,000 medical shapes, and provides annotations in the form of paired data. It is therefore also a freely available repository of 3D models for extended reality (virtual reality - VR, augmented reality - AR, mixed reality - MR) and medical 3D printing. This white paper describes in detail the motivations behind MedShapeNet, the shape acquisition procedures, the use cases, as well as the usage of the online shape search portal: https://medshapenet.ikim.nrw/
Anatomy Completor: A Multi-class Completion Framework for 3D Anatomy Reconstruction
Sep 10, 2023Abstract:In this paper, we introduce a completion framework to reconstruct the geometric shapes of various anatomies, including organs, vessels and muscles. Our work targets a scenario where one or multiple anatomies are missing in the imaging data due to surgical, pathological or traumatic factors, or simply because these anatomies are not covered by image acquisition. Automatic reconstruction of the missing anatomies benefits many applications, such as organ 3D bio-printing, whole-body segmentation, animation realism, paleoradiology and forensic imaging. We propose two paradigms based on a 3D denoising auto-encoder (DAE) to solve the anatomy reconstruction problem: (i) the DAE learns a many-to-one mapping between incomplete and complete instances; (ii) the DAE learns directly a one-to-one residual mapping between the incomplete instances and the target anatomies. We apply a loss aggregation scheme that enables the DAE to learn the many-to-one mapping more effectively and further enhances the learning of the residual mapping. On top of this, we extend the DAE to a multiclass completor by assigning a unique label to each anatomy involved. We evaluate our method using a CT dataset with whole-body segmentations. Results show that our method produces reasonable anatomy reconstructions given instances with different levels of incompleteness (i.e., one or multiple random anatomies are missing). Codes and pretrained models are publicly available at https://github.com/Jianningli/medshapenet-feedback/ tree/main/anatomy-completor
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge