Constantin Seibold
Region-Normalized DPO for Medical Image Segmentation under Noisy Judges
Jan 30, 2026Abstract:While dense pixel-wise annotations remain the gold standard for medical image segmentation, they are costly to obtain and limit scalability. In contrast, many deployed systems already produce inexpensive automatic quality-control (QC) signals like model agreement, uncertainty measures, or learned mask-quality scores which can be used for further model training without additional ground-truth annotation. However, these signals can be noisy and biased, making preference-based fine-tuning susceptible to harmful updates. We study Direct Preference Optimization (DPO) for segmentation from such noisy judges using proposals generated by a supervised base segmenter trained on a small labeled set. We find that outcomes depend strongly on how preference pairs are mined: selecting the judge's top-ranked proposal can improve peak performance when the judge is reliable, but can amplify harmful errors under weaker judges. We propose Region-Normalized DPO (RN-DPO), a segmentation-aware objective which normalizes preference updates by the size of the disagreement region between masks, reducing the leverage of harmful comparisons and improving optimization stability. Across two medical datasets and multiple regimes, RN-DPO improves sustained performance and stabilizes preference-based fine-tuning, outperforming standard DPO and strong baselines without requiring additional pixel annotations.
CT-GRAPH: Hierarchical Graph Attention Network for Anatomy-Guided CT Report Generation
Aug 07, 2025Abstract:As medical imaging is central to diagnostic processes, automating the generation of radiology reports has become increasingly relevant to assist radiologists with their heavy workloads. Most current methods rely solely on global image features, failing to capture fine-grained organ relationships crucial for accurate reporting. To this end, we propose CT-GRAPH, a hierarchical graph attention network that explicitly models radiological knowledge by structuring anatomical regions into a graph, linking fine-grained organ features to coarser anatomical systems and a global patient context. Our method leverages pretrained 3D medical feature encoders to obtain global and organ-level features by utilizing anatomical masks. These features are further refined within the graph and then integrated into a large language model to generate detailed medical reports. We evaluate our approach for the task of report generation on the large-scale chest CT dataset CT-RATE. We provide an in-depth analysis of pretrained feature encoders for CT report generation and show that our method achieves a substantial improvement of absolute 7.9\% in F1 score over current state-of-the-art methods. The code is publicly available at https://github.com/hakal104/CT-GRAPH.
Automatic Fine-grained Segmentation-assisted Report Generation
Jul 22, 2025Abstract:Reliable end-to-end clinical report generation has been a longstanding goal of medical ML research. The end goal for this process is to alleviate radiologists' workloads and provide second opinions to clinicians or patients. Thus, a necessary prerequisite for report generation models is a strong general performance and some type of innate grounding capability, to convince clinicians or patients of the veracity of the generated reports. In this paper, we present ASaRG (\textbf{A}utomatic \textbf{S}egmentation-\textbf{a}ssisted \textbf{R}eport \textbf{G}eneration), an extension of the popular LLaVA architecture that aims to tackle both of these problems. ASaRG proposes to fuse intermediate features and fine-grained segmentation maps created by specialist radiological models into LLaVA's multi-modal projection layer via simple concatenation. With a small number of added parameters, our approach achieves a +0.89\% performance gain ($p=0.012$) in CE F1 score compared to the LLaVA baseline when using only intermediate features, and +2.77\% performance gain ($p<0.001$) when adding a combination of intermediate features and fine-grained segmentation maps. Compared with COMG and ORID, two other report generation methods that utilize segmentations, the performance gain amounts to 6.98\% and 6.28\% in F1 score, respectively. ASaRG is not mutually exclusive with other changes made to the LLaVA architecture, potentially allowing our method to be combined with other advances in the field. Finally, the use of an arbitrary number of segmentations as part of the input demonstrably allows tracing elements of the report to the corresponding segmentation maps and verifying the groundedness of assessments. Our code will be made publicly available at a later date.
Is Visual in-Context Learning for Compositional Medical Tasks within Reach?
Jul 02, 2025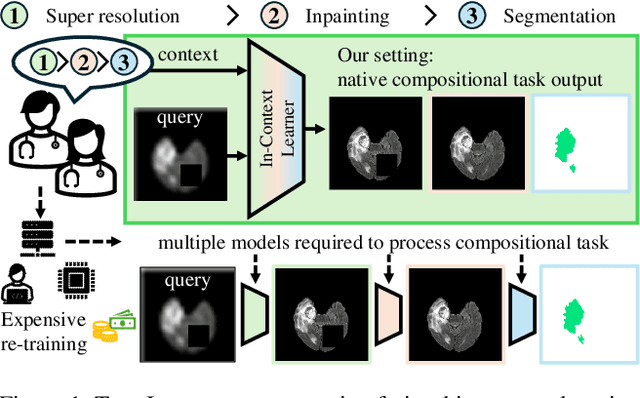
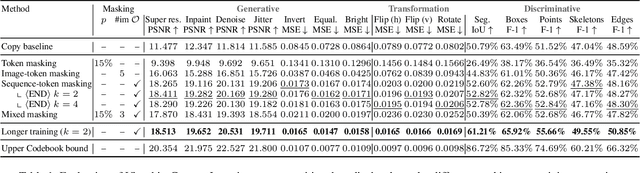
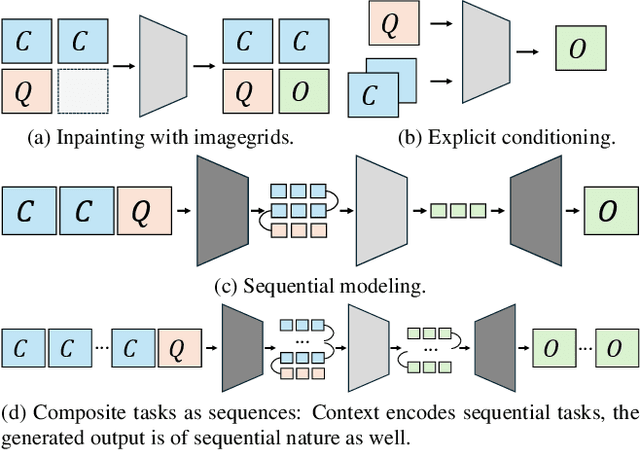
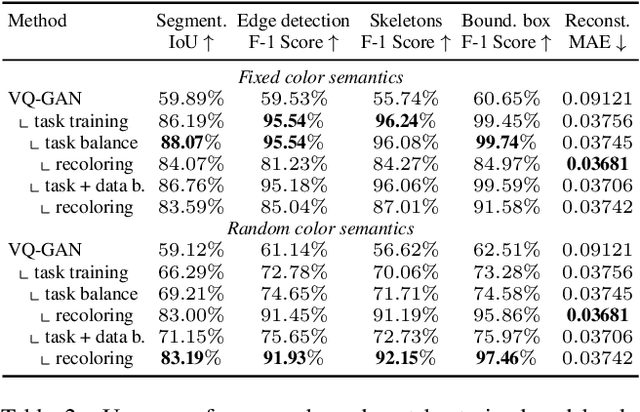
Abstract:In this paper, we explore the potential of visual in-context learning to enable a single model to handle multiple tasks and adapt to new tasks during test time without re-training. Unlike previous approaches, our focus is on training in-context learners to adapt to sequences of tasks, rather than individual tasks. Our goal is to solve complex tasks that involve multiple intermediate steps using a single model, allowing users to define entire vision pipelines flexibly at test time. To achieve this, we first examine the properties and limitations of visual in-context learning architectures, with a particular focus on the role of codebooks. We then introduce a novel method for training in-context learners using a synthetic compositional task generation engine. This engine bootstraps task sequences from arbitrary segmentation datasets, enabling the training of visual in-context learners for compositional tasks. Additionally, we investigate different masking-based training objectives to gather insights into how to train models better for solving complex, compositional tasks. Our exploration not only provides important insights especially for multi-modal medical task sequences but also highlights challenges that need to be addressed.
Good Enough: Is it Worth Improving your Label Quality?
May 27, 2025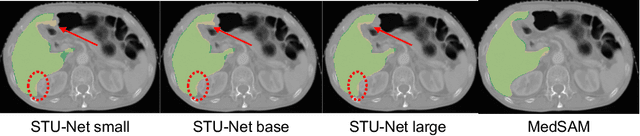
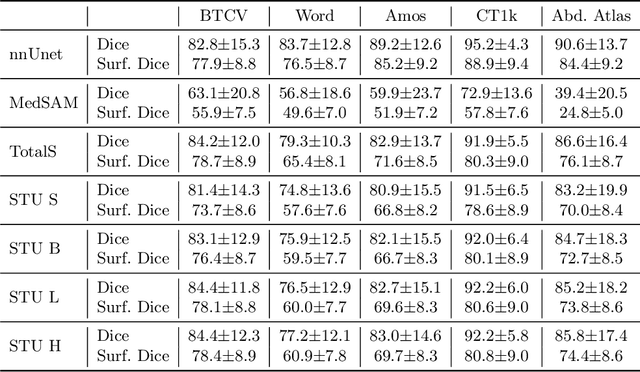
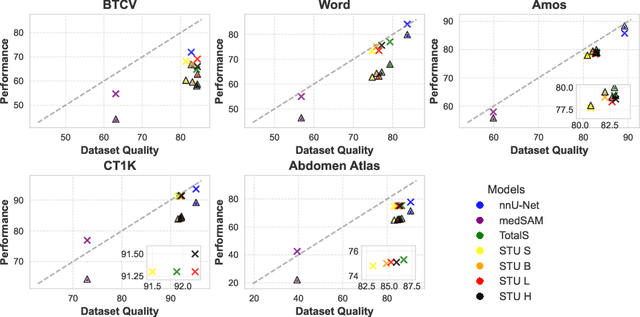
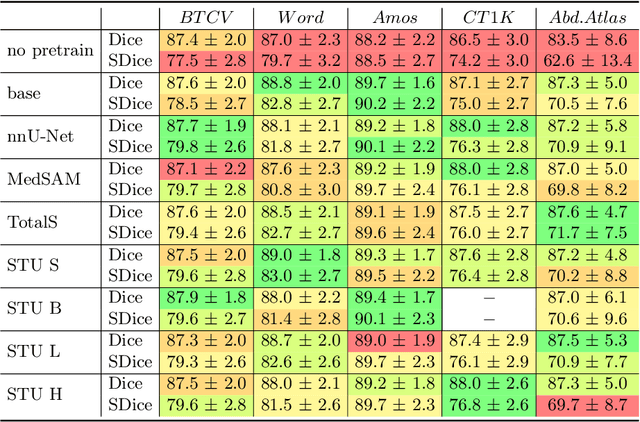
Abstract:Improving label quality in medical image segmentation is costly, but its benefits remain unclear. We systematically evaluate its impact using multiple pseudo-labeled versions of CT datasets, generated by models like nnU-Net, TotalSegmentator, and MedSAM. Our results show that while higher-quality labels improve in-domain performance, gains remain unclear if below a small threshold. For pre-training, label quality has minimal impact, suggesting that models rather transfer general concepts than detailed annotations. These findings provide guidance on when improving label quality is worth the effort.
Foreign object segmentation in chest x-rays through anatomy-guided shape insertion
Jan 21, 2025



Abstract:In this paper, we tackle the challenge of instance segmentation for foreign objects in chest radiographs, commonly seen in postoperative follow-ups with stents, pacemakers, or ingested objects in children. The diversity of foreign objects complicates dense annotation, as shown in insufficient existing datasets. To address this, we propose the simple generation of synthetic data through (1) insertion of arbitrary shapes (lines, polygons, ellipses) with varying contrasts and opacities, and (2) cut-paste augmentations from a small set of semi-automatically extracted labels. These insertions are guided by anatomy labels to ensure realistic placements, such as stents appearing only in relevant vessels. Our approach enables networks to segment complex structures with minimal manually labeled data. Notably, it achieves performance comparable to fully supervised models while using 93\% fewer manual annotations.
Every Component Counts: Rethinking the Measure of Success for Medical Semantic Segmentation in Multi-Instance Segmentation Tasks
Oct 24, 2024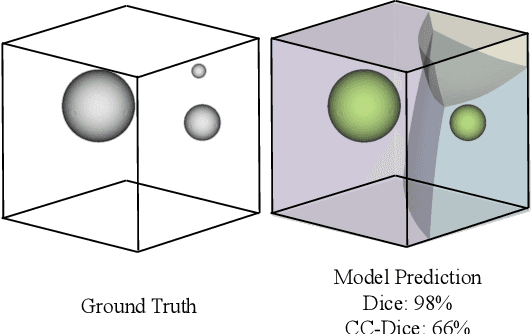
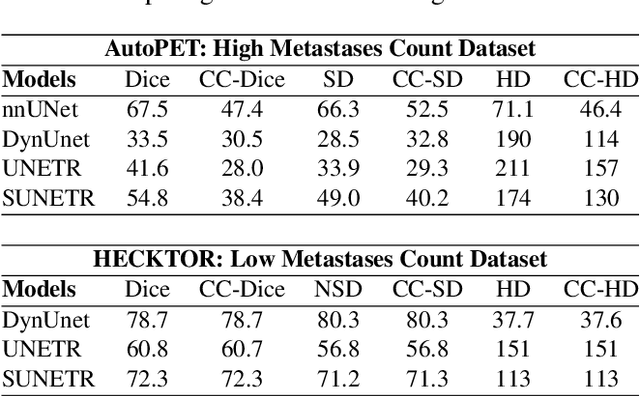
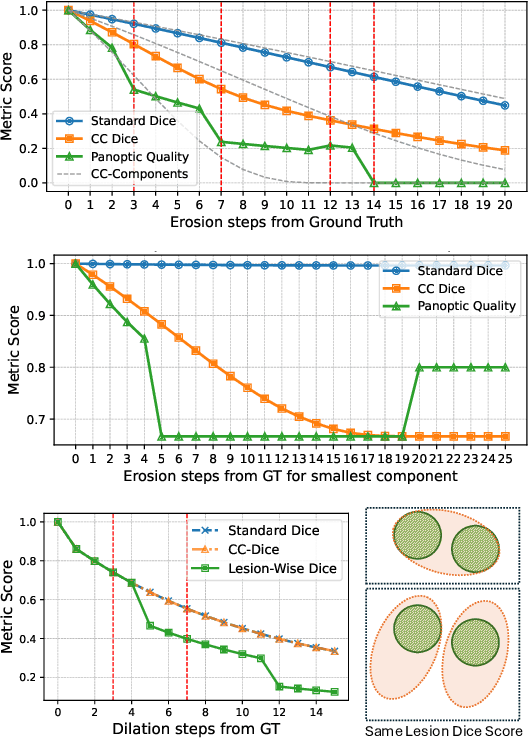
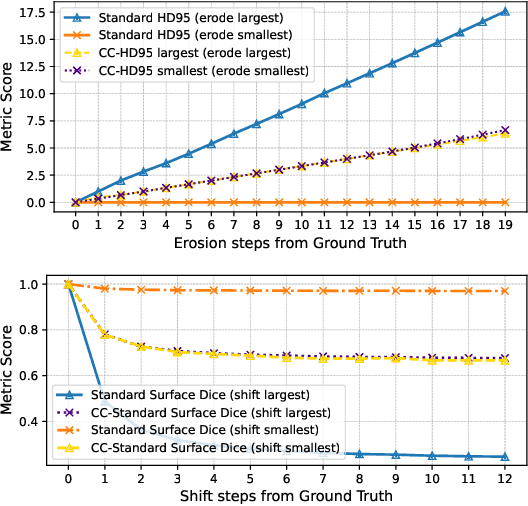
Abstract:We present Connected-Component~(CC)-Metrics, a novel semantic segmentation evaluation protocol, targeted to align existing semantic segmentation metrics to a multi-instance detection scenario in which each connected component matters. We motivate this setup in the common medical scenario of semantic metastases segmentation in a full-body PET/CT. We show how existing semantic segmentation metrics suffer from a bias towards larger connected components contradicting the clinical assessment of scans in which tumor size and clinical relevance are uncorrelated. To rebalance existing segmentation metrics, we propose to evaluate them on a per-component basis thus giving each tumor the same weight irrespective of its size. To match predictions to ground-truth segments, we employ a proximity-based matching criterion, evaluating common metrics locally at the component of interest. Using this approach, we break free of biases introduced by large metastasis for overlap-based metrics such as Dice or Surface Dice. CC-Metrics also improves distance-based metrics such as Hausdorff Distances which are uninformative for small changes that do not influence the maximum or 95th percentile, and avoids pitfalls introduced by directly combining counting-based metrics with overlap-based metrics as it is done in Panoptic Quality.
De-Identification of Medical Imaging Data: A Comprehensive Tool for Ensuring Patient Privacy
Oct 16, 2024



Abstract:Medical data employed in research frequently comprises sensitive patient health information (PHI), which is subject to rigorous legal frameworks such as the General Data Protection Regulation (GDPR) or the Health Insurance Portability and Accountability Act (HIPAA). Consequently, these types of data must be pseudonymized prior to utilisation, which presents a significant challenge for many researchers. Given the vast array of medical data, it is necessary to employ a variety of de-identification techniques. To facilitate the anonymization process for medical imaging data, we have developed an open-source tool that can be used to de-identify DICOM magnetic resonance images, computer tomography images, whole slide images and magnetic resonance twix raw data. Furthermore, the implementation of a neural network enables the removal of text within the images. The proposed tool automates an elaborate anonymization pipeline for multiple types of inputs, reducing the need for additional tools used for de-identification of imaging data. We make our code publicly available at https://github.com/code-lukas/medical_image_deidentification.
Spacewalker: Traversing Representation Spaces for Fast Interactive Exploration and Annotation of Unstructured Data
Sep 25, 2024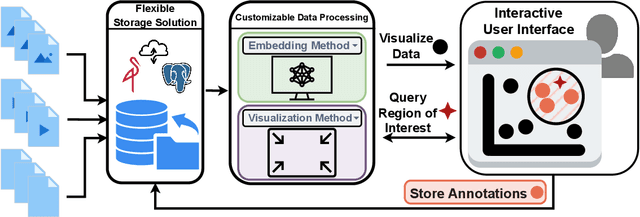
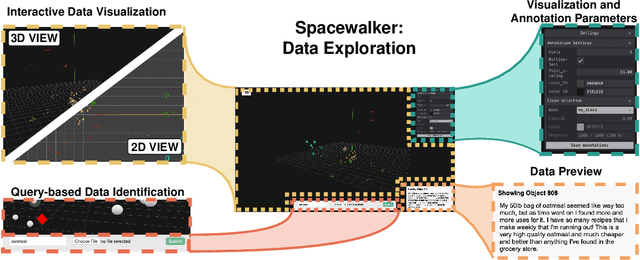
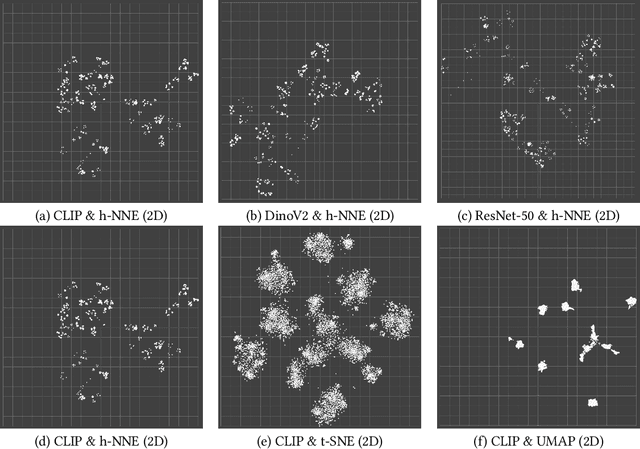
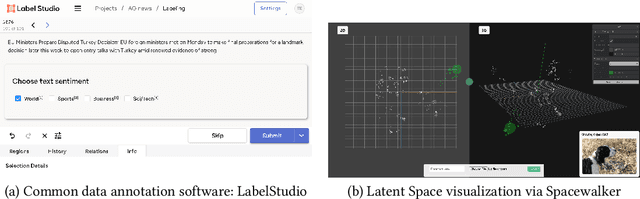
Abstract:Unstructured data in industries such as healthcare, finance, and manufacturing presents significant challenges for efficient analysis and decision making. Detecting patterns within this data and understanding their impact is critical but complex without the right tools. Traditionally, these tasks relied on the expertise of data analysts or labor-intensive manual reviews. In response, we introduce Spacewalker, an interactive tool designed to explore and annotate data across multiple modalities. Spacewalker allows users to extract data representations and visualize them in low-dimensional spaces, enabling the detection of semantic similarities. Through extensive user studies, we assess Spacewalker's effectiveness in data annotation and integrity verification. Results show that the tool's ability to traverse latent spaces and perform multi-modal queries significantly enhances the user's capacity to quickly identify relevant data. Moreover, Spacewalker allows for annotation speed-ups far superior to conventional methods, making it a promising tool for efficiently navigating unstructured data and improving decision making processes. The code of this work is open-source and can be found at: https://github.com/code-lukas/Spacewalker
Towards Synthetic Data Generation for Improved Pain Recognition in Videos under Patient Constraints
Sep 24, 2024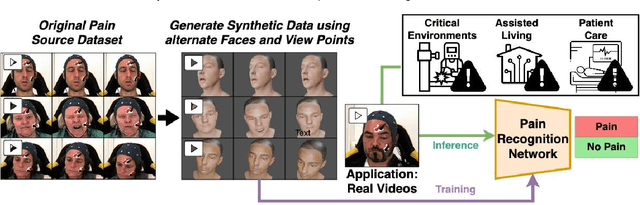
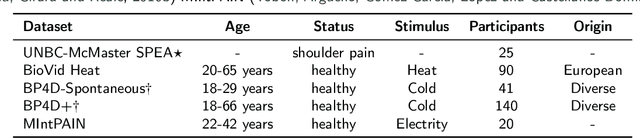
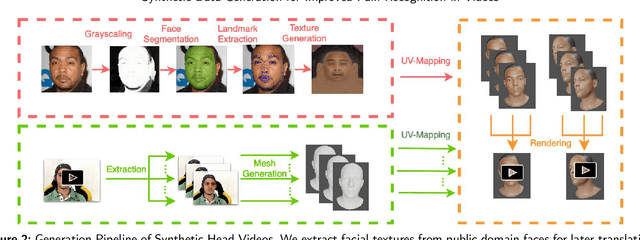
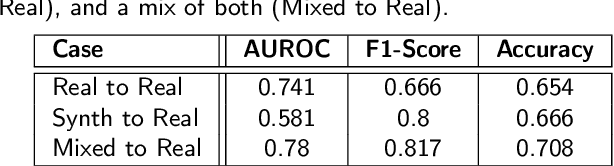
Abstract:Recognizing pain in video is crucial for improving patient-computer interaction systems, yet traditional data collection in this domain raises significant ethical and logistical challenges. This study introduces a novel approach that leverages synthetic data to enhance video-based pain recognition models, providing an ethical and scalable alternative. We present a pipeline that synthesizes realistic 3D facial models by capturing nuanced facial movements from a small participant pool, and mapping these onto diverse synthetic avatars. This process generates 8,600 synthetic faces, accurately reflecting genuine pain expressions from varied angles and perspectives. Utilizing advanced facial capture techniques, and leveraging public datasets like CelebV-HQ and FFHQ-UV for demographic diversity, our new synthetic dataset significantly enhances model training while ensuring privacy by anonymizing identities through facial replacements. Experimental results demonstrate that models trained on combinations of synthetic data paired with a small amount of real participants achieve superior performance in pain recognition, effectively bridging the gap between synthetic simulations and real-world applications. Our approach addresses data scarcity and ethical concerns, offering a new solution for pain detection and opening new avenues for research in privacy-preserving dataset generation. All resources are publicly available to encourage further innovation in this field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge