Ken Herrmann
Region-Normalized DPO for Medical Image Segmentation under Noisy Judges
Jan 30, 2026Abstract:While dense pixel-wise annotations remain the gold standard for medical image segmentation, they are costly to obtain and limit scalability. In contrast, many deployed systems already produce inexpensive automatic quality-control (QC) signals like model agreement, uncertainty measures, or learned mask-quality scores which can be used for further model training without additional ground-truth annotation. However, these signals can be noisy and biased, making preference-based fine-tuning susceptible to harmful updates. We study Direct Preference Optimization (DPO) for segmentation from such noisy judges using proposals generated by a supervised base segmenter trained on a small labeled set. We find that outcomes depend strongly on how preference pairs are mined: selecting the judge's top-ranked proposal can improve peak performance when the judge is reliable, but can amplify harmful errors under weaker judges. We propose Region-Normalized DPO (RN-DPO), a segmentation-aware objective which normalizes preference updates by the size of the disagreement region between masks, reducing the leverage of harmful comparisons and improving optimization stability. Across two medical datasets and multiple regimes, RN-DPO improves sustained performance and stabilizes preference-based fine-tuning, outperforming standard DPO and strong baselines without requiring additional pixel annotations.
CT-GRAPH: Hierarchical Graph Attention Network for Anatomy-Guided CT Report Generation
Aug 07, 2025Abstract:As medical imaging is central to diagnostic processes, automating the generation of radiology reports has become increasingly relevant to assist radiologists with their heavy workloads. Most current methods rely solely on global image features, failing to capture fine-grained organ relationships crucial for accurate reporting. To this end, we propose CT-GRAPH, a hierarchical graph attention network that explicitly models radiological knowledge by structuring anatomical regions into a graph, linking fine-grained organ features to coarser anatomical systems and a global patient context. Our method leverages pretrained 3D medical feature encoders to obtain global and organ-level features by utilizing anatomical masks. These features are further refined within the graph and then integrated into a large language model to generate detailed medical reports. We evaluate our approach for the task of report generation on the large-scale chest CT dataset CT-RATE. We provide an in-depth analysis of pretrained feature encoders for CT report generation and show that our method achieves a substantial improvement of absolute 7.9\% in F1 score over current state-of-the-art methods. The code is publicly available at https://github.com/hakal104/CT-GRAPH.
Towards Synthetic Data Generation for Improved Pain Recognition in Videos under Patient Constraints
Sep 24, 2024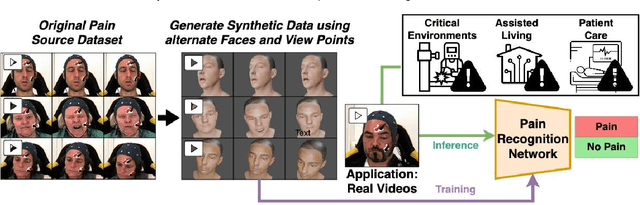
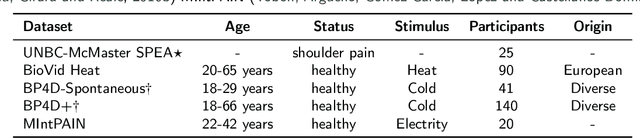
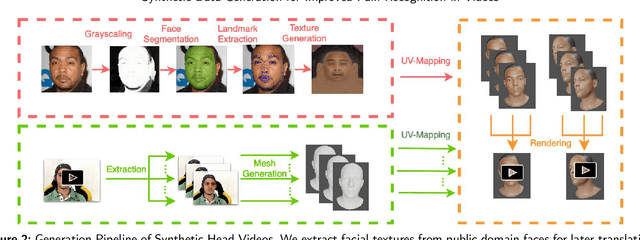
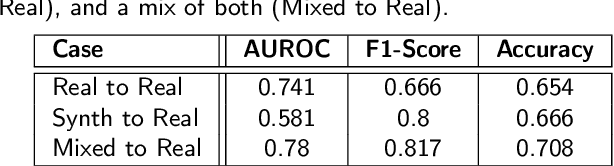
Abstract:Recognizing pain in video is crucial for improving patient-computer interaction systems, yet traditional data collection in this domain raises significant ethical and logistical challenges. This study introduces a novel approach that leverages synthetic data to enhance video-based pain recognition models, providing an ethical and scalable alternative. We present a pipeline that synthesizes realistic 3D facial models by capturing nuanced facial movements from a small participant pool, and mapping these onto diverse synthetic avatars. This process generates 8,600 synthetic faces, accurately reflecting genuine pain expressions from varied angles and perspectives. Utilizing advanced facial capture techniques, and leveraging public datasets like CelebV-HQ and FFHQ-UV for demographic diversity, our new synthetic dataset significantly enhances model training while ensuring privacy by anonymizing identities through facial replacements. Experimental results demonstrate that models trained on combinations of synthetic data paired with a small amount of real participants achieve superior performance in pain recognition, effectively bridging the gap between synthetic simulations and real-world applications. Our approach addresses data scarcity and ethical concerns, offering a new solution for pain detection and opening new avenues for research in privacy-preserving dataset generation. All resources are publicly available to encourage further innovation in this field.
Autopet III challenge: Incorporating anatomical knowledge into nnUNet for lesion segmentation in PET/CT
Sep 18, 2024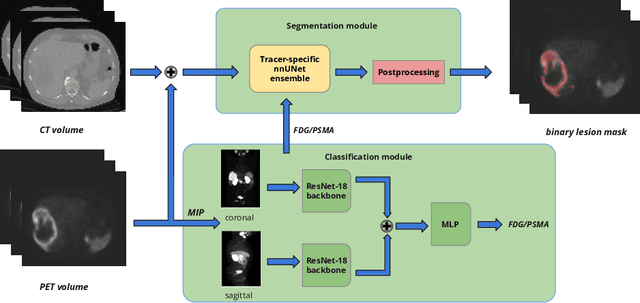
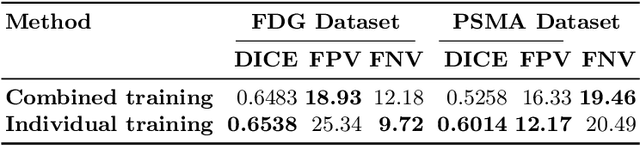
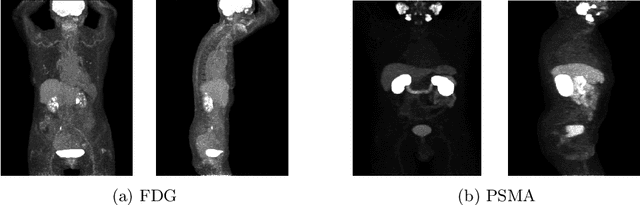

Abstract:Lesion segmentation in PET/CT imaging is essential for precise tumor characterization, which supports personalized treatment planning and enhances diagnostic precision in oncology. However, accurate manual segmentation of lesions is time-consuming and prone to inter-observer variability. Given the rising demand and clinical use of PET/CT, automated segmentation methods, particularly deep-learning-based approaches, have become increasingly more relevant. The autoPET III Challenge focuses on advancing automated segmentation of tumor lesions in PET/CT images in a multitracer multicenter setting, addressing the clinical need for quantitative, robust, and generalizable solutions. Building on previous challenges, the third iteration of the autoPET challenge introduces a more diverse dataset featuring two different tracers (FDG and PSMA) from two clinical centers. To this extent, we developed a classifier that identifies the tracer of the given PET/CT based on the Maximum Intensity Projection of the PET scan. We trained two individual nnUNet-ensembles for each tracer where anatomical labels are included as a multi-label task to enhance the model's performance. Our final submission achieves cross-validation Dice scores of 76.90% and 61.33% for the publicly available FDG and PSMA datasets, respectively. The code is available at https://github.com/hakal104/autoPETIII/ .
Anatomy-guided Pathology Segmentation
Jul 08, 2024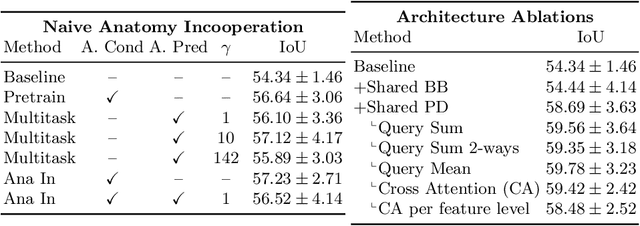
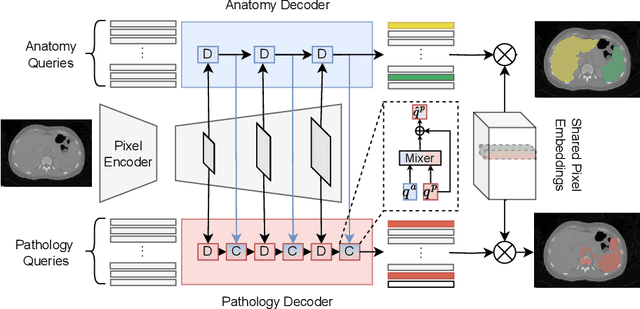
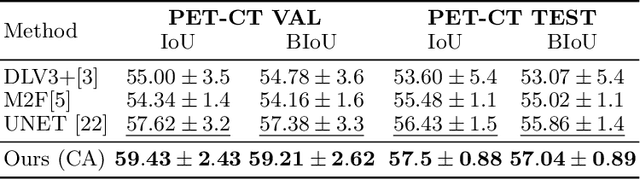
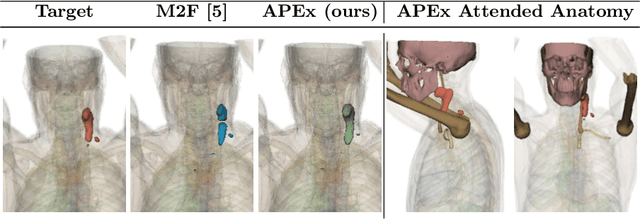
Abstract:Pathological structures in medical images are typically deviations from the expected anatomy of a patient. While clinicians consider this interplay between anatomy and pathology, recent deep learning algorithms specialize in recognizing either one of the two, rarely considering the patient's body from such a joint perspective. In this paper, we develop a generalist segmentation model that combines anatomical and pathological information, aiming to enhance the segmentation accuracy of pathological features. Our Anatomy-Pathology Exchange (APEx) training utilizes a query-based segmentation transformer which decodes a joint feature space into query-representations for human anatomy and interleaves them via a mixing strategy into the pathology-decoder for anatomy-informed pathology predictions. In doing so, we are able to report the best results across the board on FDG-PET-CT and Chest X-Ray pathology segmentation tasks with a margin of up to 3.3% as compared to strong baseline methods. Code and models will be publicly available at github.com/alexanderjaus/APEx.
Accurate Fine-Grained Segmentation of Human Anatomy in Radiographs via Volumetric Pseudo-Labeling
Jun 06, 2023Abstract:Purpose: Interpreting chest radiographs (CXR) remains challenging due to the ambiguity of overlapping structures such as the lungs, heart, and bones. To address this issue, we propose a novel method for extracting fine-grained anatomical structures in CXR using pseudo-labeling of three-dimensional computed tomography (CT) scans. Methods: We created a large-scale dataset of 10,021 thoracic CTs with 157 labels and applied an ensemble of 3D anatomy segmentation models to extract anatomical pseudo-labels. These labels were projected onto a two-dimensional plane, similar to the CXR, allowing the training of detailed semantic segmentation models for CXR without any manual annotation effort. Results: Our resulting segmentation models demonstrated remarkable performance on CXR, with a high average model-annotator agreement between two radiologists with mIoU scores of 0.93 and 0.85 for frontal and lateral anatomy, while inter-annotator agreement remained at 0.95 and 0.83 mIoU. Our anatomical segmentations allowed for the accurate extraction of relevant explainable medical features such as the cardio-thoracic-ratio. Conclusion: Our method of volumetric pseudo-labeling paired with CT projection offers a promising approach for detailed anatomical segmentation of CXR with a high agreement with human annotators. This technique may have important clinical implications, particularly in the analysis of various thoracic pathologies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge