Moon Kim
LIMIS: Towards Language-based Interactive Medical Image Segmentation
Oct 22, 2024Abstract:Within this work, we introduce LIMIS: The first purely language-based interactive medical image segmentation model. We achieve this by adapting Grounded SAM to the medical domain and designing a language-based model interaction strategy that allows radiologists to incorporate their knowledge into the segmentation process. LIMIS produces high-quality initial segmentation masks by leveraging medical foundation models and allows users to adapt segmentation masks using only language, opening up interactive segmentation to scenarios where physicians require using their hands for other tasks. We evaluate LIMIS on three publicly available medical datasets in terms of performance and usability with experts from the medical domain confirming its high-quality segmentation masks and its interactive usability.
ReXamine-Global: A Framework for Uncovering Inconsistencies in Radiology Report Generation Metrics
Aug 29, 2024Abstract:Given the rapidly expanding capabilities of generative AI models for radiology, there is a need for robust metrics that can accurately measure the quality of AI-generated radiology reports across diverse hospitals. We develop ReXamine-Global, a LLM-powered, multi-site framework that tests metrics across different writing styles and patient populations, exposing gaps in their generalization. First, our method tests whether a metric is undesirably sensitive to reporting style, providing different scores depending on whether AI-generated reports are stylistically similar to ground-truth reports or not. Second, our method measures whether a metric reliably agrees with experts, or whether metric and expert scores of AI-generated report quality diverge for some sites. Using 240 reports from 6 hospitals around the world, we apply ReXamine-Global to 7 established report evaluation metrics and uncover serious gaps in their generalizability. Developers can apply ReXamine-Global when designing new report evaluation metrics, ensuring their robustness across sites. Additionally, our analysis of existing metrics can guide users of those metrics towards evaluation procedures that work reliably at their sites of interest.
Anatomy-guided Pathology Segmentation
Jul 08, 2024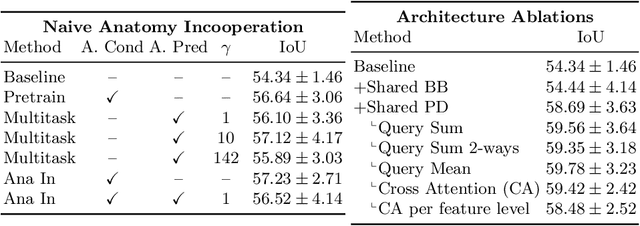
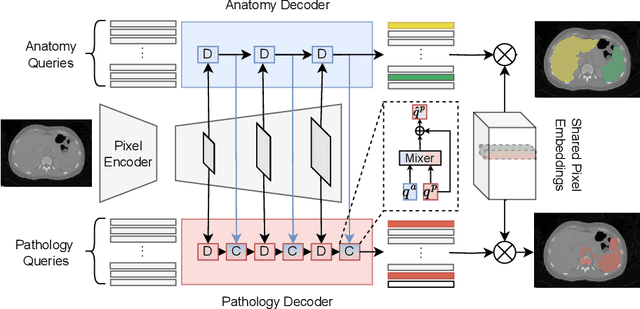
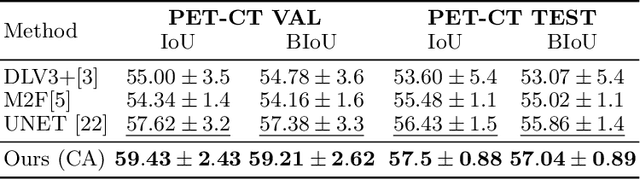
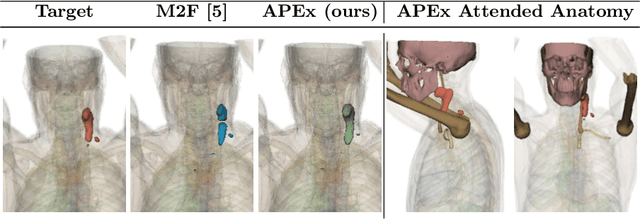
Abstract:Pathological structures in medical images are typically deviations from the expected anatomy of a patient. While clinicians consider this interplay between anatomy and pathology, recent deep learning algorithms specialize in recognizing either one of the two, rarely considering the patient's body from such a joint perspective. In this paper, we develop a generalist segmentation model that combines anatomical and pathological information, aiming to enhance the segmentation accuracy of pathological features. Our Anatomy-Pathology Exchange (APEx) training utilizes a query-based segmentation transformer which decodes a joint feature space into query-representations for human anatomy and interleaves them via a mixing strategy into the pathology-decoder for anatomy-informed pathology predictions. In doing so, we are able to report the best results across the board on FDG-PET-CT and Chest X-Ray pathology segmentation tasks with a margin of up to 3.3% as compared to strong baseline methods. Code and models will be publicly available at github.com/alexanderjaus/APEx.
Real-World Federated Learning in Radiology: Hurdles to overcome and Benefits to gain
May 15, 2024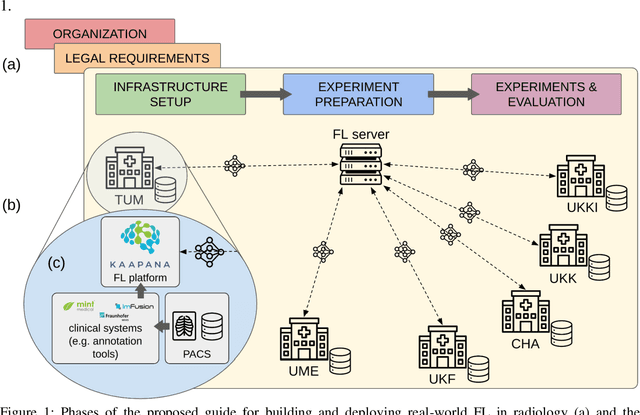



Abstract:Objective: Federated Learning (FL) enables collaborative model training while keeping data locally. Currently, most FL studies in radiology are conducted in simulated environments due to numerous hurdles impeding its translation into practice. The few existing real-world FL initiatives rarely communicate specific measures taken to overcome these hurdles, leaving behind a significant knowledge gap. Minding efforts to implement real-world FL, there is a notable lack of comprehensive assessment comparing FL to less complex alternatives. Materials & Methods: We extensively reviewed FL literature, categorizing insights along with our findings according to their nature and phase while establishing a FL initiative, summarized to a comprehensive guide. We developed our own FL infrastructure within the German Radiological Cooperative Network (RACOON) and demonstrated its functionality by training FL models on lung pathology segmentation tasks across six university hospitals. We extensively evaluated FL against less complex alternatives in three distinct evaluation scenarios. Results: The proposed guide outlines essential steps, identified hurdles, and proposed solutions for establishing successful FL initiatives conducting real-world experiments. Our experimental results show that FL outperforms less complex alternatives in all evaluation scenarios, justifying the effort required to translate FL into real-world applications. Discussion & Conclusion: Our proposed guide aims to aid future FL researchers in circumventing pitfalls and accelerating translation of FL into radiological applications. Our results underscore the value of efforts needed to translate FL into real-world applications by demonstrating advantageous performance over alternatives, and emphasize the importance of strategic organization, robust management of distributed data and infrastructure in real-world settings.
Rethinking Annotator Simulation: Realistic Evaluation of Whole-Body PET Lesion Interactive Segmentation Methods
Apr 02, 2024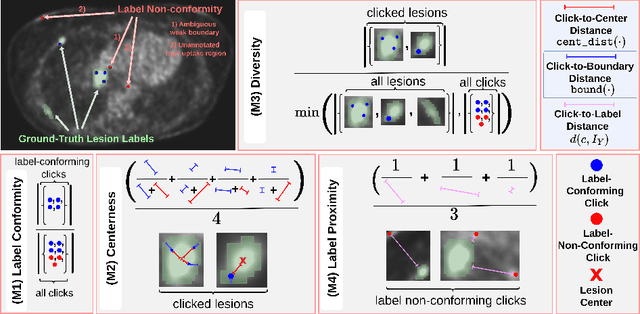
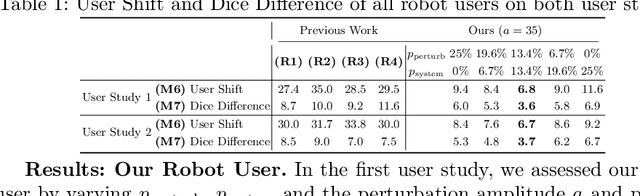
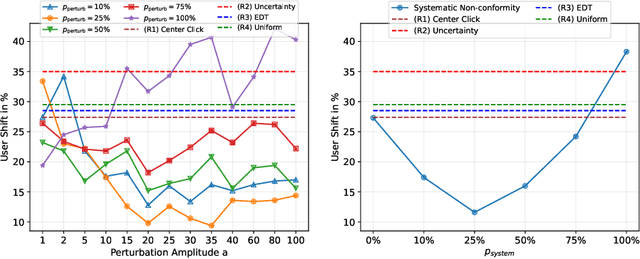
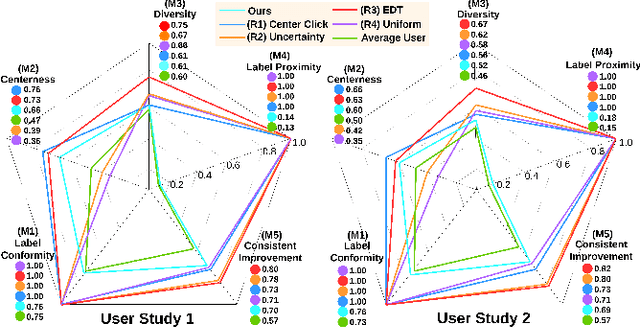
Abstract:Interactive segmentation plays a crucial role in accelerating the annotation, particularly in domains requiring specialized expertise such as nuclear medicine. For example, annotating lesions in whole-body Positron Emission Tomography (PET) images can require over an hour per volume. While previous works evaluate interactive segmentation models through either real user studies or simulated annotators, both approaches present challenges. Real user studies are expensive and often limited in scale, while simulated annotators, also known as robot users, tend to overestimate model performance due to their idealized nature. To address these limitations, we introduce four evaluation metrics that quantify the user shift between real and simulated annotators. In an initial user study involving four annotators, we assess existing robot users using our proposed metrics and find that robot users significantly deviate in performance and annotation behavior compared to real annotators. Based on these findings, we propose a more realistic robot user that reduces the user shift by incorporating human factors such as click variation and inter-annotator disagreement. We validate our robot user in a second user study, involving four other annotators, and show it consistently reduces the simulated-to-real user shift compared to traditional robot users. By employing our robot user, we can conduct more large-scale and cost-efficient evaluations of interactive segmentation models, while preserving the fidelity of real user studies. Our implementation is based on MONAI Label and will be made publicly available.
Sliding Window FastEdit: A Framework for Lesion Annotation in Whole-body PET Images
Nov 24, 2023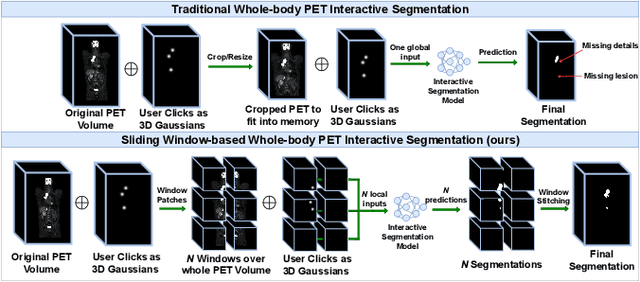
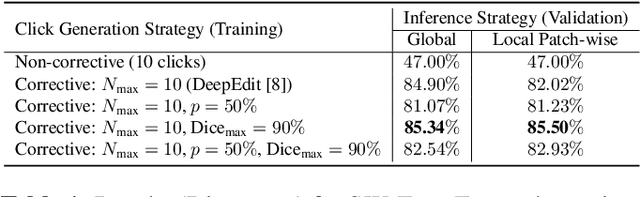
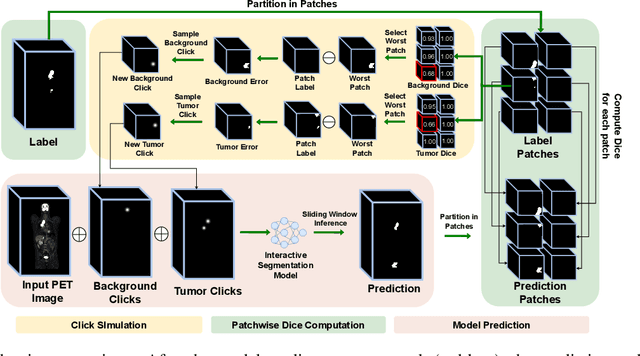
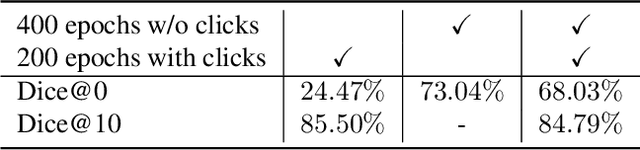
Abstract:Deep learning has revolutionized the accurate segmentation of diseases in medical imaging. However, achieving such results requires training with numerous manual voxel annotations. This requirement presents a challenge for whole-body Positron Emission Tomography (PET) imaging, where lesions are scattered throughout the body. To tackle this problem, we introduce SW-FastEdit - an interactive segmentation framework that accelerates the labeling by utilizing only a few user clicks instead of voxelwise annotations. While prior interactive models crop or resize PET volumes due to memory constraints, we use the complete volume with our sliding window-based interactive scheme. Our model outperforms existing non-sliding window interactive models on the AutoPET dataset and generalizes to the previously unseen HECKTOR dataset. A user study revealed that annotators achieve high-quality predictions with only 10 click iterations and a low perceived NASA-TLX workload. Our framework is implemented using MONAI Label and is available: https://github.com/matt3o/AutoPET2-Submission/
Multilingual Natural Language Processing Model for Radiology Reports -- The Summary is all you need!
Oct 06, 2023



Abstract:The impression section of a radiology report summarizes important radiology findings and plays a critical role in communicating these findings to physicians. However, the preparation of these summaries is time-consuming and error-prone for radiologists. Recently, numerous models for radiology report summarization have been developed. Nevertheless, there is currently no model that can summarize these reports in multiple languages. Such a model could greatly improve future research and the development of Deep Learning models that incorporate data from patients with different ethnic backgrounds. In this study, the generation of radiology impressions in different languages was automated by fine-tuning a model, publicly available, based on a multilingual text-to-text Transformer to summarize findings available in English, Portuguese, and German radiology reports. In a blind test, two board-certified radiologists indicated that for at least 70% of the system-generated summaries, the quality matched or exceeded the corresponding human-written summaries, suggesting substantial clinical reliability. Furthermore, this study showed that the multilingual model outperformed other models that specialized in summarizing radiology reports in only one language, as well as models that were not specifically designed for summarizing radiology reports, such as ChatGPT.
Why does my medical AI look at pictures of birds? Exploring the efficacy of transfer learning across domain boundaries
Jun 30, 2023Abstract:It is an open secret that ImageNet is treated as the panacea of pretraining. Particularly in medical machine learning, models not trained from scratch are often finetuned based on ImageNet-pretrained models. We posit that pretraining on data from the domain of the downstream task should almost always be preferred instead. We leverage RadNet-12M, a dataset containing more than 12 million computed tomography (CT) image slices, to explore the efficacy of self-supervised pretraining on medical and natural images. Our experiments cover intra- and cross-domain transfer scenarios, varying data scales, finetuning vs. linear evaluation, and feature space analysis. We observe that intra-domain transfer compares favorably to cross-domain transfer, achieving comparable or improved performance (0.44% - 2.07% performance increase using RadNet pretraining, depending on the experiment) and demonstrate the existence of a domain boundary-related generalization gap and domain-specific learned features.
Accurate Fine-Grained Segmentation of Human Anatomy in Radiographs via Volumetric Pseudo-Labeling
Jun 06, 2023Abstract:Purpose: Interpreting chest radiographs (CXR) remains challenging due to the ambiguity of overlapping structures such as the lungs, heart, and bones. To address this issue, we propose a novel method for extracting fine-grained anatomical structures in CXR using pseudo-labeling of three-dimensional computed tomography (CT) scans. Methods: We created a large-scale dataset of 10,021 thoracic CTs with 157 labels and applied an ensemble of 3D anatomy segmentation models to extract anatomical pseudo-labels. These labels were projected onto a two-dimensional plane, similar to the CXR, allowing the training of detailed semantic segmentation models for CXR without any manual annotation effort. Results: Our resulting segmentation models demonstrated remarkable performance on CXR, with a high average model-annotator agreement between two radiologists with mIoU scores of 0.93 and 0.85 for frontal and lateral anatomy, while inter-annotator agreement remained at 0.95 and 0.83 mIoU. Our anatomical segmentations allowed for the accurate extraction of relevant explainable medical features such as the cardio-thoracic-ratio. Conclusion: Our method of volumetric pseudo-labeling paired with CT projection offers a promising approach for detailed anatomical segmentation of CXR with a high agreement with human annotators. This technique may have important clinical implications, particularly in the analysis of various thoracic pathologies.
Multimodal Interactive Lung Lesion Segmentation: A Framework for Annotating PET/CT Images based on Physiological and Anatomical Cues
Jan 24, 2023



Abstract:Recently, deep learning enabled the accurate segmentation of various diseases in medical imaging. These performances, however, typically demand large amounts of manual voxel annotations. This tedious process for volumetric data becomes more complex when not all required information is available in a single imaging domain as is the case for PET/CT data. We propose a multimodal interactive segmentation framework that mitigates these issues by combining anatomical and physiological cues from PET/CT data. Our framework utilizes the geodesic distance transform to represent the user annotations and we implement a novel ellipsoid-based user simulation scheme during training. We further propose two annotation interfaces and conduct a user study to estimate their usability. We evaluated our model on the in-domain validation dataset and an unseen PET/CT dataset. We make our code publicly available: https://github.com/verena-hallitschke/pet-ct-annotate.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge