Frederic Jonske
Region-Normalized DPO for Medical Image Segmentation under Noisy Judges
Jan 30, 2026Abstract:While dense pixel-wise annotations remain the gold standard for medical image segmentation, they are costly to obtain and limit scalability. In contrast, many deployed systems already produce inexpensive automatic quality-control (QC) signals like model agreement, uncertainty measures, or learned mask-quality scores which can be used for further model training without additional ground-truth annotation. However, these signals can be noisy and biased, making preference-based fine-tuning susceptible to harmful updates. We study Direct Preference Optimization (DPO) for segmentation from such noisy judges using proposals generated by a supervised base segmenter trained on a small labeled set. We find that outcomes depend strongly on how preference pairs are mined: selecting the judge's top-ranked proposal can improve peak performance when the judge is reliable, but can amplify harmful errors under weaker judges. We propose Region-Normalized DPO (RN-DPO), a segmentation-aware objective which normalizes preference updates by the size of the disagreement region between masks, reducing the leverage of harmful comparisons and improving optimization stability. Across two medical datasets and multiple regimes, RN-DPO improves sustained performance and stabilizes preference-based fine-tuning, outperforming standard DPO and strong baselines without requiring additional pixel annotations.
Automatic Fine-grained Segmentation-assisted Report Generation
Jul 22, 2025Abstract:Reliable end-to-end clinical report generation has been a longstanding goal of medical ML research. The end goal for this process is to alleviate radiologists' workloads and provide second opinions to clinicians or patients. Thus, a necessary prerequisite for report generation models is a strong general performance and some type of innate grounding capability, to convince clinicians or patients of the veracity of the generated reports. In this paper, we present ASaRG (\textbf{A}utomatic \textbf{S}egmentation-\textbf{a}ssisted \textbf{R}eport \textbf{G}eneration), an extension of the popular LLaVA architecture that aims to tackle both of these problems. ASaRG proposes to fuse intermediate features and fine-grained segmentation maps created by specialist radiological models into LLaVA's multi-modal projection layer via simple concatenation. With a small number of added parameters, our approach achieves a +0.89\% performance gain ($p=0.012$) in CE F1 score compared to the LLaVA baseline when using only intermediate features, and +2.77\% performance gain ($p<0.001$) when adding a combination of intermediate features and fine-grained segmentation maps. Compared with COMG and ORID, two other report generation methods that utilize segmentations, the performance gain amounts to 6.98\% and 6.28\% in F1 score, respectively. ASaRG is not mutually exclusive with other changes made to the LLaVA architecture, potentially allowing our method to be combined with other advances in the field. Finally, the use of an arbitrary number of segmentations as part of the input demonstrably allows tracing elements of the report to the corresponding segmentation maps and verifying the groundedness of assessments. Our code will be made publicly available at a later date.
ReXamine-Global: A Framework for Uncovering Inconsistencies in Radiology Report Generation Metrics
Aug 29, 2024Abstract:Given the rapidly expanding capabilities of generative AI models for radiology, there is a need for robust metrics that can accurately measure the quality of AI-generated radiology reports across diverse hospitals. We develop ReXamine-Global, a LLM-powered, multi-site framework that tests metrics across different writing styles and patient populations, exposing gaps in their generalization. First, our method tests whether a metric is undesirably sensitive to reporting style, providing different scores depending on whether AI-generated reports are stylistically similar to ground-truth reports or not. Second, our method measures whether a metric reliably agrees with experts, or whether metric and expert scores of AI-generated report quality diverge for some sites. Using 240 reports from 6 hospitals around the world, we apply ReXamine-Global to 7 established report evaluation metrics and uncover serious gaps in their generalizability. Developers can apply ReXamine-Global when designing new report evaluation metrics, ensuring their robustness across sites. Additionally, our analysis of existing metrics can guide users of those metrics towards evaluation procedures that work reliably at their sites of interest.
MedShapeNet -- A Large-Scale Dataset of 3D Medical Shapes for Computer Vision
Sep 12, 2023



Abstract:We present MedShapeNet, a large collection of anatomical shapes (e.g., bones, organs, vessels) and 3D surgical instrument models. Prior to the deep learning era, the broad application of statistical shape models (SSMs) in medical image analysis is evidence that shapes have been commonly used to describe medical data. Nowadays, however, state-of-the-art (SOTA) deep learning algorithms in medical imaging are predominantly voxel-based. In computer vision, on the contrary, shapes (including, voxel occupancy grids, meshes, point clouds and implicit surface models) are preferred data representations in 3D, as seen from the numerous shape-related publications in premier vision conferences, such as the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), as well as the increasing popularity of ShapeNet (about 51,300 models) and Princeton ModelNet (127,915 models) in computer vision research. MedShapeNet is created as an alternative to these commonly used shape benchmarks to facilitate the translation of data-driven vision algorithms to medical applications, and it extends the opportunities to adapt SOTA vision algorithms to solve critical medical problems. Besides, the majority of the medical shapes in MedShapeNet are modeled directly on the imaging data of real patients, and therefore it complements well existing shape benchmarks comprising of computer-aided design (CAD) models. MedShapeNet currently includes more than 100,000 medical shapes, and provides annotations in the form of paired data. It is therefore also a freely available repository of 3D models for extended reality (virtual reality - VR, augmented reality - AR, mixed reality - MR) and medical 3D printing. This white paper describes in detail the motivations behind MedShapeNet, the shape acquisition procedures, the use cases, as well as the usage of the online shape search portal: https://medshapenet.ikim.nrw/
Why does my medical AI look at pictures of birds? Exploring the efficacy of transfer learning across domain boundaries
Jun 30, 2023Abstract:It is an open secret that ImageNet is treated as the panacea of pretraining. Particularly in medical machine learning, models not trained from scratch are often finetuned based on ImageNet-pretrained models. We posit that pretraining on data from the domain of the downstream task should almost always be preferred instead. We leverage RadNet-12M, a dataset containing more than 12 million computed tomography (CT) image slices, to explore the efficacy of self-supervised pretraining on medical and natural images. Our experiments cover intra- and cross-domain transfer scenarios, varying data scales, finetuning vs. linear evaluation, and feature space analysis. We observe that intra-domain transfer compares favorably to cross-domain transfer, achieving comparable or improved performance (0.44% - 2.07% performance increase using RadNet pretraining, depending on the experiment) and demonstrate the existence of a domain boundary-related generalization gap and domain-specific learned features.
MOMO -- Deep Learning-driven classification of external DICOM studies for PACS archivation
Dec 01, 2021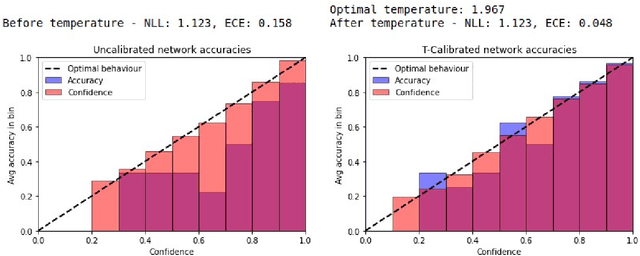
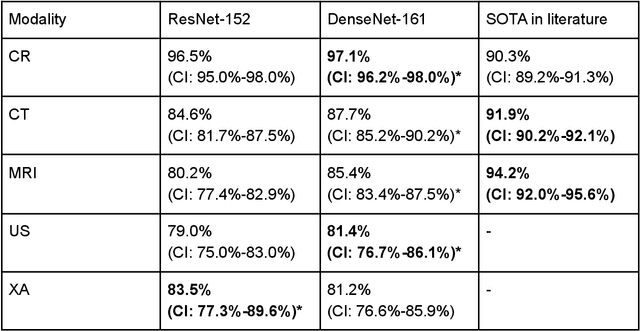
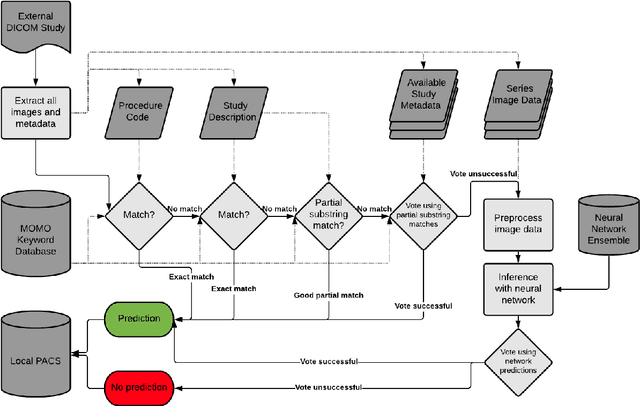
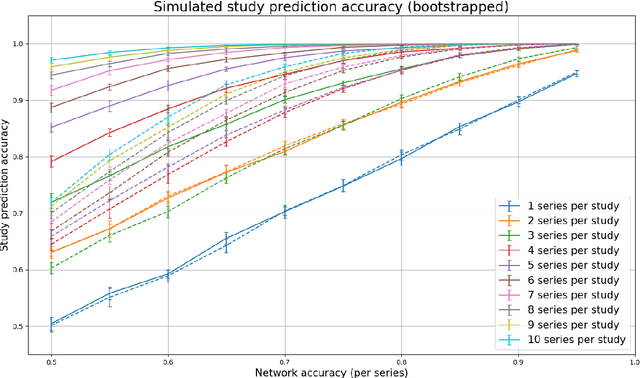
Abstract:Patients regularly continue assessment or treatment in other facilities than they began them in, receiving their previous imaging studies as a CD-ROM and requiring clinical staff at the new hospital to import these studies into their local database. However, between different facilities, standards for nomenclature, contents, or even medical procedures may vary, often requiring human intervention to accurately classify the received studies in the context of the recipient hospital's standards. In this study, the authors present MOMO (MOdality Mapping and Orchestration), a deep learning-based approach to automate this mapping process utilizing metadata substring matching and a neural network ensemble, which is trained to recognize the 76 most common imaging studies across seven different modalities. A retrospective study is performed to measure the accuracy that this algorithm can provide. To this end, a set of 11,934 imaging series with existing labels was retrieved from the local hospital's PACS database to train the neural networks. A set of 843 completely anonymized external studies was hand-labeled to assess the performance of our algorithm. Additionally, an ablation study was performed to measure the performance impact of the network ensemble in the algorithm, and a comparative performance test with a commercial product was conducted. In comparison to a commercial product (96.20% predictive power, 82.86% accuracy, 1.36% minor errors), a neural network ensemble alone performs the classification task with less accuracy (99.05% predictive power, 72.69% accuracy, 10.3% minor errors). However, MOMO outperforms either by a large margin in accuracy and with increased predictive power (99.29% predictive power, 92.71% accuracy, 2.63% minor errors).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge