Johannes Haubold
Princeton University
Efficient Complex-Valued Vision Transformers for MRI Classification Directly from k-Space
Jan 26, 2026Abstract:Deep learning applications in Magnetic Resonance Imaging (MRI) predominantly operate on reconstructed magnitude images, a process that discards phase information and requires computationally expensive transforms. Standard neural network architectures rely on local operations (convolutions or grid-patches) that are ill-suited for the global, non-local nature of raw frequency-domain (k-Space) data. In this work, we propose a novel complex-valued Vision Transformer (kViT) designed to perform classification directly on k-Space data. To bridge the geometric disconnect between current architectures and MRI physics, we introduce a radial k-Space patching strategy that respects the spectral energy distribution of the frequency-domain. Extensive experiments on the fastMRI and in-house datasets demonstrate that our approach achieves classification performance competitive with state-of-the-art image-domain baselines (ResNet, EfficientNet, ViT). Crucially, kViT exhibits superior robustness to high acceleration factors and offers a paradigm shift in computational efficiency, reducing VRAM consumption during training by up to 68$\times$ compared to standard methods. This establishes a pathway for resource-efficient, direct-from-scanner AI analysis.
An Annotated Dataset of Errors in Premodern Greek and Baselines for Detecting Them
Oct 14, 2024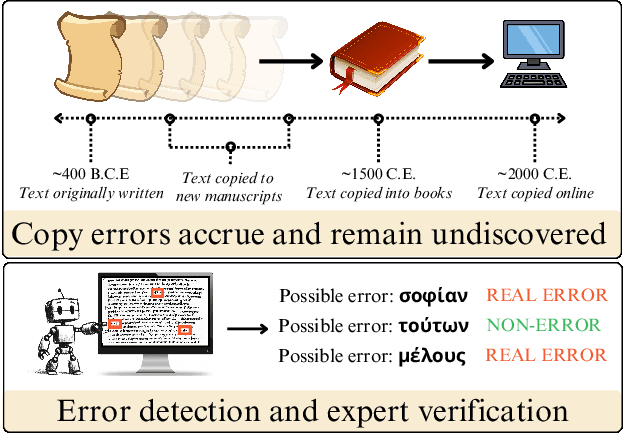
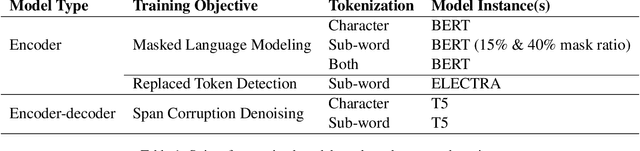
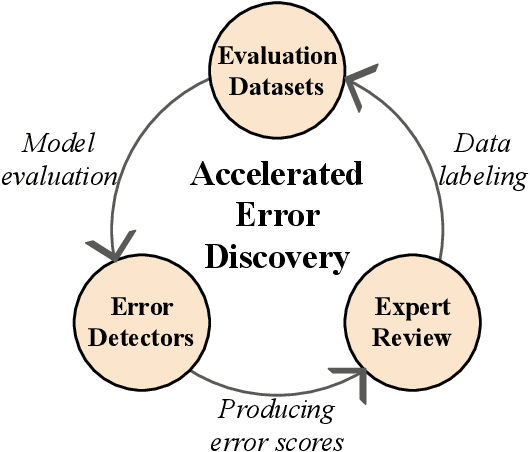
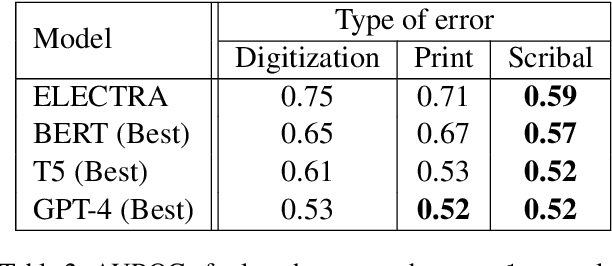
Abstract:As premodern texts are passed down over centuries, errors inevitably accrue. These errors can be challenging to identify, as some have survived undetected for so long precisely because they are so elusive. While prior work has evaluated error detection methods on artificially-generated errors, we introduce the first dataset of real errors in premodern Greek, enabling the evaluation of error detection methods on errors that genuinely accumulated at some stage in the centuries-long copying process. To create this dataset, we use metrics derived from BERT conditionals to sample 1,000 words more likely to contain errors, which are then annotated and labeled by a domain expert as errors or not. We then propose and evaluate new error detection methods and find that our discriminator-based detector outperforms all other methods, improving the true positive rate for classifying real errors by 5%. We additionally observe that scribal errors are more difficult to detect than print or digitization errors. Our dataset enables the evaluation of error detection methods on real errors in premodern texts for the first time, providing a benchmark for developing more effective error detection algorithms to assist scholars in restoring premodern works.
ReXamine-Global: A Framework for Uncovering Inconsistencies in Radiology Report Generation Metrics
Aug 29, 2024Abstract:Given the rapidly expanding capabilities of generative AI models for radiology, there is a need for robust metrics that can accurately measure the quality of AI-generated radiology reports across diverse hospitals. We develop ReXamine-Global, a LLM-powered, multi-site framework that tests metrics across different writing styles and patient populations, exposing gaps in their generalization. First, our method tests whether a metric is undesirably sensitive to reporting style, providing different scores depending on whether AI-generated reports are stylistically similar to ground-truth reports or not. Second, our method measures whether a metric reliably agrees with experts, or whether metric and expert scores of AI-generated report quality diverge for some sites. Using 240 reports from 6 hospitals around the world, we apply ReXamine-Global to 7 established report evaluation metrics and uncover serious gaps in their generalizability. Developers can apply ReXamine-Global when designing new report evaluation metrics, ensuring their robustness across sites. Additionally, our analysis of existing metrics can guide users of those metrics towards evaluation procedures that work reliably at their sites of interest.
SALT: Introducing a Framework for Hierarchical Segmentations in Medical Imaging using Softmax for Arbitrary Label Trees
Jul 11, 2024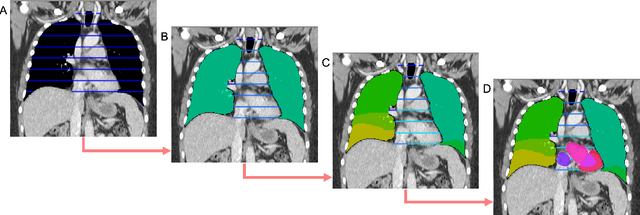
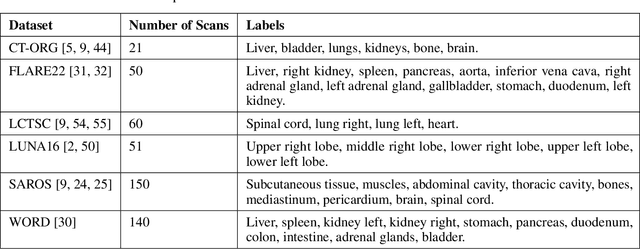
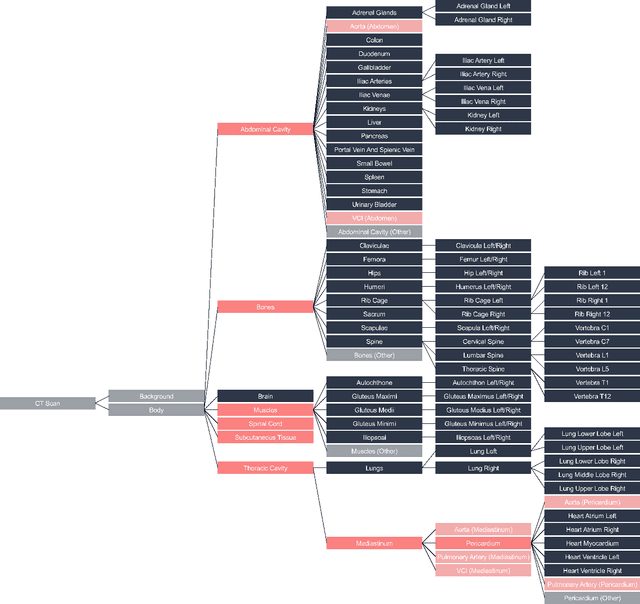
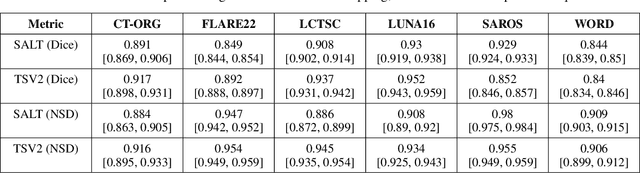
Abstract:Traditional segmentation networks approach anatomical structures as standalone elements, overlooking the intrinsic hierarchical connections among them. This study introduces Softmax for Arbitrary Label Trees (SALT), a novel approach designed to leverage the hierarchical relationships between labels, improving the efficiency and interpretability of the segmentations. This study introduces a novel segmentation technique for CT imaging, which leverages conditional probabilities to map the hierarchical structure of anatomical landmarks, such as the spine's division into lumbar, thoracic, and cervical regions and further into individual vertebrae. The model was developed using the SAROS dataset from The Cancer Imaging Archive (TCIA), comprising 900 body region segmentations from 883 patients. The dataset was further enhanced by generating additional segmentations with the TotalSegmentator, for a total of 113 labels. The model was trained on 600 scans, while validation and testing were conducted on 150 CT scans. Performance was assessed using the Dice score across various datasets, including SAROS, CT-ORG, FLARE22, LCTSC, LUNA16, and WORD. Among the evaluated datasets, SALT achieved its best results on the LUNA16 and SAROS datasets, with Dice scores of 0.93 and 0.929 respectively. The model demonstrated reliable accuracy across other datasets, scoring 0.891 on CT-ORG and 0.849 on FLARE22. The LCTSC dataset showed a score of 0.908 and the WORD dataset also showed good performance with a score of 0.844. SALT used the hierarchical structures inherent in the human body to achieve whole-body segmentations with an average of 35 seconds for 100 slices. This rapid processing underscores its potential for integration into clinical workflows, facilitating the automatic and efficient computation of full-body segmentations with each CT scan, thus enhancing diagnostic processes and patient care.
Gadolinium dose reduction for brain MRI using conditional deep learning
Mar 06, 2024



Abstract:Recently, deep learning (DL)-based methods have been proposed for the computational reduction of gadolinium-based contrast agents (GBCAs) to mitigate adverse side effects while preserving diagnostic value. Currently, the two main challenges for these approaches are the accurate prediction of contrast enhancement and the synthesis of realistic images. In this work, we address both challenges by utilizing the contrast signal encoded in the subtraction images of pre-contrast and post-contrast image pairs. To avoid the synthesis of any noise or artifacts and solely focus on contrast signal extraction and enhancement from low-dose subtraction images, we train our DL model using noise-free standard-dose subtraction images as targets. As a result, our model predicts the contrast enhancement signal only; thereby enabling synthesization of images beyond the standard dose. Furthermore, we adapt the embedding idea of recent diffusion-based models to condition our model on physical parameters affecting the contrast enhancement behavior. We demonstrate the effectiveness of our approach on synthetic and real datasets using various scanners, field strengths, and contrast agents.
Towards Unifying Anatomy Segmentation: Automated Generation of a Full-body CT Dataset via Knowledge Aggregation and Anatomical Guidelines
Jul 25, 2023Abstract:In this study, we present a method for generating automated anatomy segmentation datasets using a sequential process that involves nnU-Net-based pseudo-labeling and anatomy-guided pseudo-label refinement. By combining various fragmented knowledge bases, we generate a dataset of whole-body CT scans with $142$ voxel-level labels for 533 volumes providing comprehensive anatomical coverage which experts have approved. Our proposed procedure does not rely on manual annotation during the label aggregation stage. We examine its plausibility and usefulness using three complementary checks: Human expert evaluation which approved the dataset, a Deep Learning usefulness benchmark on the BTCV dataset in which we achieve 85% dice score without using its training dataset, and medical validity checks. This evaluation procedure combines scalable automated checks with labor-intensive high-quality expert checks. Besides the dataset, we release our trained unified anatomical segmentation model capable of predicting $142$ anatomical structures on CT data.
Why does my medical AI look at pictures of birds? Exploring the efficacy of transfer learning across domain boundaries
Jun 30, 2023Abstract:It is an open secret that ImageNet is treated as the panacea of pretraining. Particularly in medical machine learning, models not trained from scratch are often finetuned based on ImageNet-pretrained models. We posit that pretraining on data from the domain of the downstream task should almost always be preferred instead. We leverage RadNet-12M, a dataset containing more than 12 million computed tomography (CT) image slices, to explore the efficacy of self-supervised pretraining on medical and natural images. Our experiments cover intra- and cross-domain transfer scenarios, varying data scales, finetuning vs. linear evaluation, and feature space analysis. We observe that intra-domain transfer compares favorably to cross-domain transfer, achieving comparable or improved performance (0.44% - 2.07% performance increase using RadNet pretraining, depending on the experiment) and demonstrate the existence of a domain boundary-related generalization gap and domain-specific learned features.
Logion: Machine Learning for Greek Philology
May 01, 2023Abstract:This paper presents machine-learning methods to address various problems in Greek philology. After training a BERT model on the largest premodern Greek dataset used for this purpose to date, we identify and correct previously undetected errors made by scribes in the process of textual transmission, in what is, to our knowledge, the first successful identification of such errors via machine learning. Additionally, we demonstrate the model's capacity to fill gaps caused by material deterioration of premodern manuscripts and compare the model's performance to that of a domain expert. We find that best performance is achieved when the domain expert is provided with model suggestions for inspiration. With such human-computer collaborations in mind, we explore the model's interpretability and find that certain attention heads appear to encode select grammatical features of premodern Greek.
k-strip: A novel segmentation algorithm in k-space for the application of skull stripping
May 19, 2022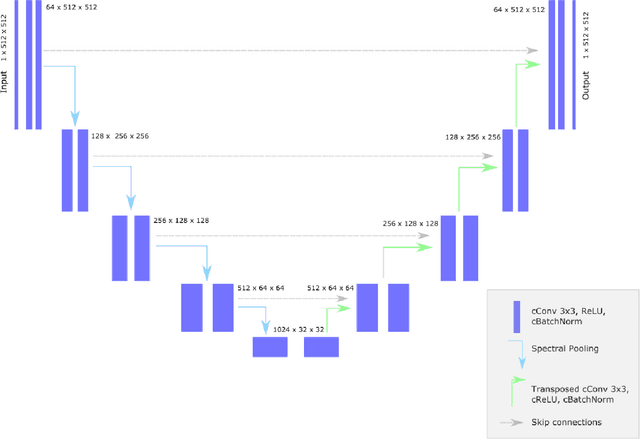
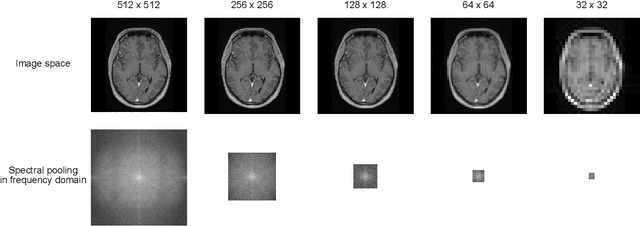
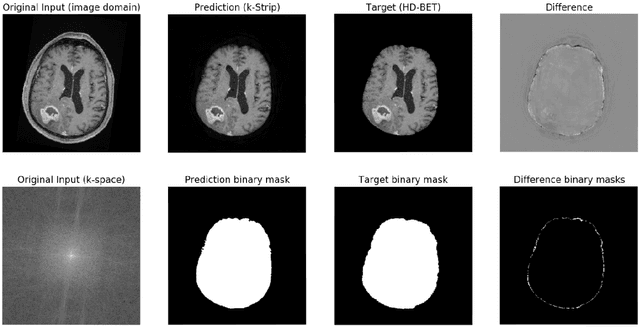
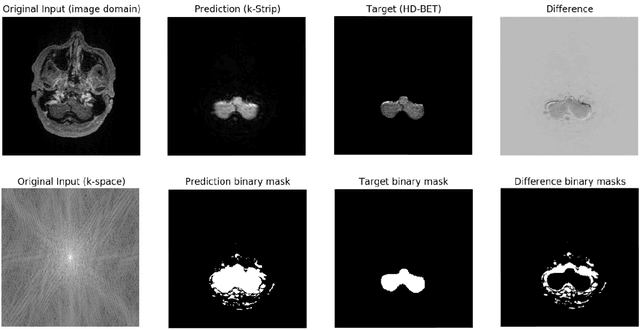
Abstract:Objectives: Present a novel deep learning-based skull stripping algorithm for magnetic resonance imaging (MRI) that works directly in the information rich k-space. Materials and Methods: Using two datasets from different institutions with a total of 36,900 MRI slices, we trained a deep learning-based model to work directly with the complex raw k-space data. Skull stripping performed by HD-BET (Brain Extraction Tool) in the image domain were used as the ground truth. Results: Both datasets were very similar to the ground truth (DICE scores of 92\%-98\% and Hausdorff distances of under 5.5 mm). Results on slices above the eye-region reach DICE scores of up to 99\%, while the accuracy drops in regions around the eyes and below, with partially blurred output. The output of k-strip often smoothed edges at the demarcation to the skull. Binary masks are created with an appropriate threshold. Conclusion: With this proof-of-concept study, we were able to show the feasibility of working in the k-space frequency domain, preserving phase information, with consistent results. Future research should be dedicated to discovering additional ways the k-space can be used for innovative image analysis and further workflows.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge