Kelsey L. Pomykala
'A net for everyone': fully personalized and unsupervised neural networks trained with longitudinal data from a single patient
Oct 25, 2022Abstract:With the rise in importance of personalized medicine, we trained personalized neural networks to detect tumor progression in longitudinal datasets. The model was evaluated on two datasets with a total of 64 scans from 32 patients diagnosed with glioblastoma multiforme (GBM). Contrast-enhanced T1w sequences of brain magnetic resonance imaging (MRI) images were used in this study. For each patient, we trained their own neural network using just two images from different timepoints. Our approach uses a Wasserstein-GAN (generative adversarial network), an unsupervised network architecture, to map the differences between the two images. Using this map, the change in tumor volume can be evaluated. Due to the combination of data augmentation and the network architecture, co-registration of the two images is not needed. Furthermore, we do not rely on any additional training data, (manual) annotations or pre-training neural networks. The model received an AUC-score of 0.87 for tumor change. We also introduced a modified RANO criteria, for which an accuracy of 66% can be achieved. We show that using data from just one patient can be used to train deep neural networks to monitor tumor change.
GAN-based generation of realistic 3D data: A systematic review and taxonomy
Jul 04, 2022



Abstract:Data has become the most valuable resource in today's world. With the massive proliferation of data-driven algorithms, such as deep learning-based approaches, the availability of data is of great interest. In this context, high-quality training, validation and testing datasets are particularly needed. Volumetric data is a very important resource in medicine, as it ranges from disease diagnoses to therapy monitoring. When the dataset is sufficient, models can be trained to help doctors with these tasks. Unfortunately, there are scenarios and applications where large amounts of data is unavailable. For example, in the medical field, rare diseases and privacy issues can lead to restricted data availability. In non-medical fields, the high cost of obtaining a sufficient amount of high-quality data can also be a concern. A solution to these problems can be the generation of synthetic data to perform data augmentation in combination with other more traditional methods of data augmentation. Therefore, most of the publications on 3D Generative Adversarial Networks (GANs) are within the medical domain. The existence of mechanisms to generate realistic synthetic data is a good asset to overcome this challenge, especially in healthcare, as the data must be of good quality and close to reality, i.e. realistic, and without privacy issues. In this review, we provide a summary of works that generate realistic 3D synthetic data using GANs. We therefore outline GAN-based methods in these areas with common architectures, advantages and disadvantages. We present a novel taxonomy, evaluations, challenges and research opportunities to provide a holistic overview of the current state of GANs in medicine and other fields.
k-strip: A novel segmentation algorithm in k-space for the application of skull stripping
May 19, 2022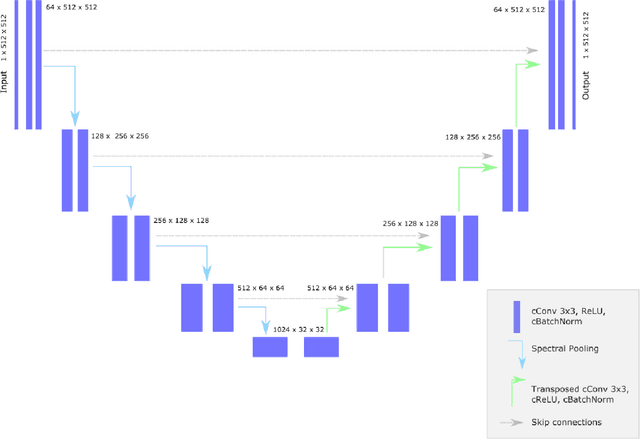
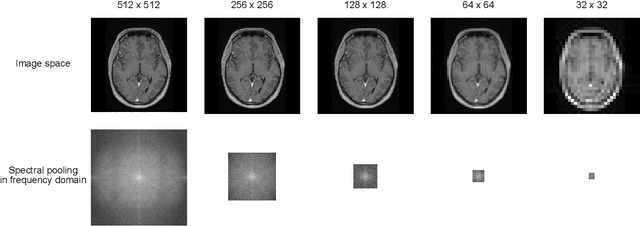
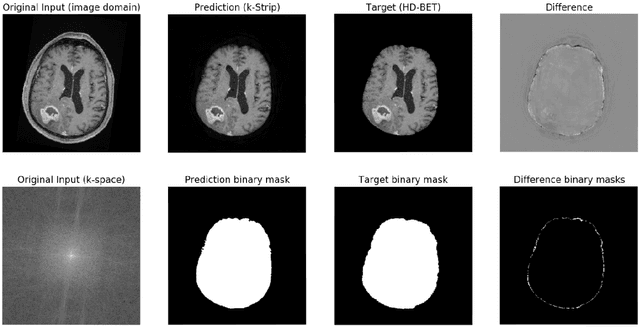
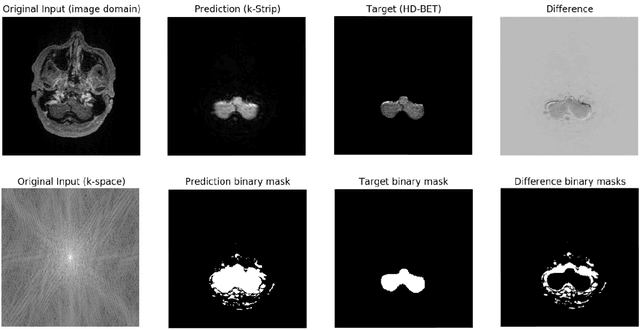
Abstract:Objectives: Present a novel deep learning-based skull stripping algorithm for magnetic resonance imaging (MRI) that works directly in the information rich k-space. Materials and Methods: Using two datasets from different institutions with a total of 36,900 MRI slices, we trained a deep learning-based model to work directly with the complex raw k-space data. Skull stripping performed by HD-BET (Brain Extraction Tool) in the image domain were used as the ground truth. Results: Both datasets were very similar to the ground truth (DICE scores of 92\%-98\% and Hausdorff distances of under 5.5 mm). Results on slices above the eye-region reach DICE scores of up to 99\%, while the accuracy drops in regions around the eyes and below, with partially blurred output. The output of k-strip often smoothed edges at the demarcation to the skull. Binary masks are created with an appropriate threshold. Conclusion: With this proof-of-concept study, we were able to show the feasibility of working in the k-space frequency domain, preserving phase information, with consistent results. Future research should be dedicated to discovering additional ways the k-space can be used for innovative image analysis and further workflows.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge