Victor Alves
Centro Algoritmi, University of Minho, Braga, Portugal
Curated endoscopic retrograde cholangiopancreatography images dataset
Jan 23, 2026Abstract:Endoscopic Retrograde Cholangiopancreatography (ERCP) is a key procedure in the diagnosis and treatment of biliary and pancreatic diseases. Artificial intelligence has been pointed as one solution to automatize diagnosis. However, public ERCP datasets are scarce, which limits the use of such approach. Therefore, this study aims to help fill this gap by providing a large and curated dataset. The collection is composed of 19.018 raw images and 19.317 processed from 1.602 patients. 5.519 images are labeled, which provides a ready to use dataset. All images were manually inspected and annotated by two gastroenterologist with more than 5 years of experience and reviewed by another gastroenterologist with more than 20 years of experience, all with more than 400 ERCP procedures annually. The utility and validity of the dataset is proven by a classification experiment. This collection aims to provide or contribute for a benchmark in automatic ERCP analysis and diagnosis of biliary and pancreatic diseases.
Radiomics and Clinical Features in Predictive Modelling of Brain Metastases Recurrence
Dec 17, 2025



Abstract:Brain metastases affect approximately between 20% and 40% of cancer patients and are commonly treated with radiotherapy or radiosurgery. Early prediction of recurrence following treatment could enable timely clinical intervention and improve patient outcomes. This study proposes an artificial intelligence based approach for predicting brain metastasis recurrence using multimodal imaging and clinical data. A retrospective cohort of 97 patients was collected, including Computed Tomography (CT) and Magnetic Resonance Imaging (MRI) acquired before treatment and at first follow-up, together with relevant clinical variables. Image preprocessing included CT windowing and artifact reduction, MRI enhancement, and multimodal CT MRI registration. After applying inclusion criteria, 53 patients were retained for analysis. Radiomics features were extracted from the imaging data, and delta radiomics was employed to characterize temporal changes between pre-treatment and follow-up scans. Multiple machine learning classifiers were trained and evaluated, including an analysis of discrepancies between treatment planning target volumes and delivered isodose volumes. Despite limitations related to sample size and class imbalance, the results demonstrate the feasibility of radiomics based models, namely ensemble models, for recurrence prediction and suggest a potential association between radiation dose discrepancies and recurrence risk. This work supports further investigation of AI-driven tools to assist clinical decision-making in brain metastasis management.
Enhancing Privacy: The Utility of Stand-Alone Synthetic CT and MRI for Tumor and Bone Segmentation
Jun 13, 2025Abstract:AI requires extensive datasets, while medical data is subject to high data protection. Anonymization is essential, but poses a challenge for some regions, such as the head, as identifying structures overlap with regions of clinical interest. Synthetic data offers a potential solution, but studies often lack rigorous evaluation of realism and utility. Therefore, we investigate to what extent synthetic data can replace real data in segmentation tasks. We employed head and neck cancer CT scans and brain glioma MRI scans from two large datasets. Synthetic data were generated using generative adversarial networks and diffusion models. We evaluated the quality of the synthetic data using MAE, MS-SSIM, Radiomics and a Visual Turing Test (VTT) performed by 5 radiologists and their usefulness in segmentation tasks using DSC. Radiomics indicates high fidelity of synthetic MRIs, but fall short in producing highly realistic CT tissue, with correlation coefficient of 0.8784 and 0.5461 for MRI and CT tumors, respectively. DSC results indicate limited utility of synthetic data: tumor segmentation achieved DSC=0.064 on CT and 0.834 on MRI, while bone segmentation a mean DSC=0.841. Relation between DSC and correlation is observed, but is limited by the complexity of the task. VTT results show synthetic CTs' utility, but with limited educational applications. Synthetic data can be used independently for the segmentation task, although limited by the complexity of the structures to segment. Advancing generative models to better tolerate heterogeneous inputs and learn subtle details is essential for enhancing their realism and expanding their application potential.
A Simultaneous Approach for Training Neural Differential-Algebraic Systems of Equations
Apr 07, 2025



Abstract:Scientific machine learning is an emerging field that broadly describes the combination of scientific computing and machine learning to address challenges in science and engineering. Within the context of differential equations, this has produced highly influential methods, such as neural ordinary differential equations (NODEs). Recent works extend this line of research to consider neural differential-algebraic systems of equations (DAEs), where some unknown relationships within the DAE are learned from data. Training neural DAEs, similarly to neural ODEs, is computationally expensive, as it requires the solution of a DAE for every parameter update. Further, the rigorous consideration of algebraic constraints is difficult within common deep learning training algorithms such as stochastic gradient descent. In this work, we apply the simultaneous approach to neural DAE problems, resulting in a fully discretized nonlinear optimization problem, which is solved to local optimality and simultaneously obtains the neural network parameters and the solution to the corresponding DAE. We extend recent work demonstrating the simultaneous approach for neural ODEs, by presenting a general framework to solve neural DAEs, with explicit consideration of hybrid models, where some components of the DAE are known, e.g. physics-informed constraints. Furthermore, we present a general strategy for improving the performance and convergence of the nonlinear programming solver, based on solving an auxiliary problem for initialization and approximating Hessian terms. We achieve promising results in terms of accuracy, model generalizability and computational cost, across different problem settings such as sparse data, unobserved states and multiple trajectories. Lastly, we provide several promising future directions to improve the scalability and robustness of our approach.
The Impact of Artificial Intelligence on Emergency Medicine: A Review of Recent Advances
Mar 17, 2025Abstract:Artificial Intelligence (AI) is revolutionizing emergency medicine by enhancing diagnostic processes and improving patient outcomes. This article provides a review of the current applications of AI in emergency imaging studies, focusing on the last five years of advancements. AI technologies, particularly machine learning and deep learning, are pivotal in interpreting complex imaging data, offering rapid, accurate diagnoses and potentially surpassing traditional diagnostic methods. Studies highlighted within the article demonstrate AI's capabilities in accurately detecting conditions such as fractures, pneumothorax, and pulmonary diseases from various imaging modalities including X-rays, CT scans, and MRIs. Furthermore, AI's ability to predict clinical outcomes like mechanical ventilation needs illustrates its potential in crisis resource optimization. Despite these advancements, the integration of AI into clinical practice presents challenges such as data privacy, algorithmic bias, and the need for extensive validation across diverse settings. This review underscores the transformative potential of AI in emergency settings, advocating for a future where AI and clinical expertise synergize to elevate patient care standards.
RFUDS -- A Brain Metastases Imaging Dataset of Radiotherapy Follow-Up
Dec 21, 2024Abstract:Brain metastases are a common diagnosis that affects between 20% and 40% of cancer patients. Subsequent to radiation therapy, patients with brain metastases undergo follow-up sessions during which the response to treatment is monitored. In this study, a dataset of medical images from 44 patients with at least one brain metastasis and different primary tumor locations was collected and processed. Each patient was treated with either a linear accelerator or a gamma knife. Computed Tomography (CT) and Magnetic Resonance Imaging (MRI) scans were collected at various time points, including before treatment and during follow-up sessions. The CT datasets were processed using windowing and artifact reduction techniques, while the MRI datasets were subjected to CLAHE. The NifTI files corresponding to the CT and MRI images were made public available. In order to align the datasets of each patient, a multimodal registration was performed between the CT and MRI datasets, with different software options being tested. The fusion matrices were provided together with the dataset. The aforementioned steps resulted in the creation of an optimized dataset, prepared for use in a range of studies related to brain metastases. RFUds is publicity available at zenodo under the DOI 10.5281/zenodo.14524784.
Comparative Analysis of nnUNet and MedNeXt for Head and Neck Tumor Segmentation in MRI-guided Radiotherapy
Nov 22, 2024Abstract:Radiation therapy (RT) is essential in treating head and neck cancer (HNC), with magnetic resonance imaging(MRI)-guided RT offering superior soft tissue contrast and functional imaging. However, manual tumor segmentation is time-consuming and complex, and therfore remains a challenge. In this study, we present our solution as team TUMOR to the HNTS-MRG24 MICCAI Challenge which is focused on automated segmentation of primary gross tumor volumes (GTVp) and metastatic lymph node gross tumor volume (GTVn) in pre-RT and mid-RT MRI images. We utilized the HNTS-MRG2024 dataset, which consists of 150 MRI scans from patients diagnosed with HNC, including original and registered pre-RT and mid-RT T2-weighted images with corresponding segmentation masks for GTVp and GTVn. We employed two state-of-the-art models in deep learning, nnUNet and MedNeXt. For Task 1, we pretrained models on pre-RT registered and mid-RT images, followed by fine-tuning on original pre-RT images. For Task 2, we combined registered pre-RT images, registered pre-RT segmentation masks, and mid-RT data as a multi-channel input for training. Our solution for Task 1 achieved 1st place in the final test phase with an aggregated Dice Similarity Coefficient of 0.8254, and our solution for Task 2 ranked 8th with a score of 0.7005. The proposed solution is publicly available at Github Repository.
Brain Tumour Removing and Missing Modality Generation using 3D WDM
Nov 07, 2024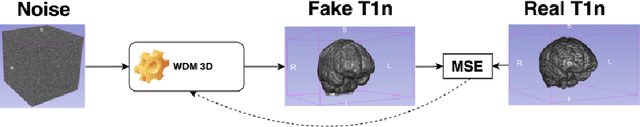
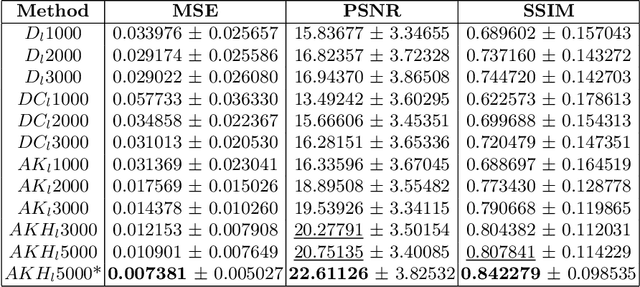
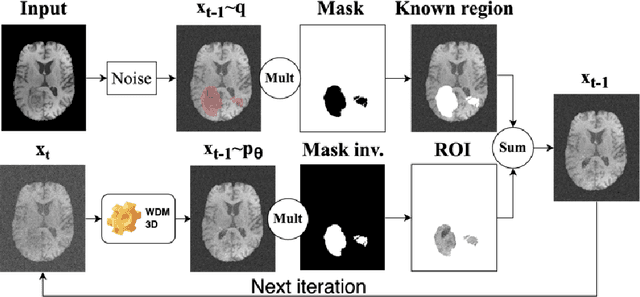
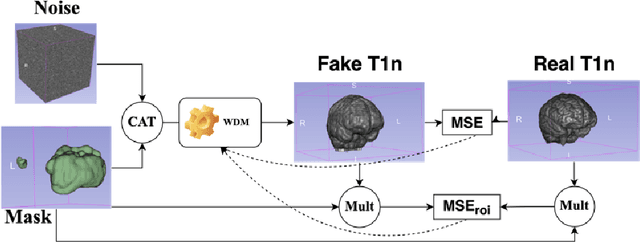
Abstract:This paper presents the second-placed solution for task 8 and the participation solution for task 7 of BraTS 2024. The adoption of automated brain analysis algorithms to support clinical practice is increasing. However, many of these algorithms struggle with the presence of brain lesions or the absence of certain MRI modalities. The alterations in the brain's morphology leads to high variability and thus poor performance of predictive models that were trained only on healthy brains. The lack of information that is usually provided by some of the missing MRI modalities also reduces the reliability of the prediction models trained with all modalities. In order to improve the performance of these models, we propose the use of conditional 3D wavelet diffusion models. The wavelet transform enabled full-resolution image training and prediction on a GPU with 48 GB VRAM, without patching or downsampling, preserving all information for prediction. For the inpainting task of BraTS 2024, the use of a large and variable number of healthy masks and the stability and efficiency of the 3D wavelet diffusion model resulted in 0.007, 22.61 and 0.842 in the validation set and 0.07 , 22.8 and 0.91 in the testing set (MSE, PSNR and SSIM respectively). The code for these tasks is available at https://github.com/ShadowTwin41/BraTS_2023_2024_solutions.
Improved Multi-Task Brain Tumour Segmentation with Synthetic Data Augmentation
Nov 07, 2024



Abstract:This paper presents the winning solution of task 1 and the third-placed solution of task 3 of the BraTS challenge. The use of automated tools in clinical practice has increased due to the development of more and more sophisticated and reliable algorithms. However, achieving clinical standards and developing tools for real-life scenarios is a major challenge. To this end, BraTS has organised tasks to find the most advanced solutions for specific purposes. In this paper, we propose the use of synthetic data to train state-of-the-art frameworks in order to improve the segmentation of adult gliomas in a post-treatment scenario, and the segmentation of meningioma for radiotherapy planning. Our results suggest that the use of synthetic data leads to more robust algorithms, although the synthetic data generation pipeline is not directly suited to the meningioma task. The code for these tasks is available at https://github.com/ShadowTwin41/BraTS_2023_2024_solutions.
Subthalamic Nucleus segmentation in high-field Magnetic Resonance data. Is space normalization by template co-registration necessary?
Jul 22, 2024Abstract:Deep Brain Stimulation (DBS) is one of the most successful methods to diminish late-stage Parkinson's Disease (PD) symptoms. It is a delicate surgical procedure which requires detailed pre-surgical patient's study. High-field Magnetic Resonance Imaging (MRI) has proven its improved capacity of capturing the Subthalamic Nucleus (STN) - the main target of DBS in PD - in greater detail than lower field images. Here, we present a comparison between the performance of two different Deep Learning (DL) automatic segmentation architectures, one based in the registration to a brain template and the other performing the segmentation in in the MRI acquisition native space. The study was based on publicly available high-field 7 Tesla (T) brain MRI datasets of T1-weighted and T2-weighted sequences. nnUNet was used on the segmentation step of both architectures, while the data pre and post-processing pipelines diverged. The evaluation metrics showed that the performance of the segmentation directly in the native space yielded better results for the STN segmentation, despite not showing any advantage over the template-based method for the to other analysed structures: the Red Nucleus (RN) and the Substantia Nigra (SN).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge