Kunpeng Xie
Enhancing Privacy: The Utility of Stand-Alone Synthetic CT and MRI for Tumor and Bone Segmentation
Jun 13, 2025Abstract:AI requires extensive datasets, while medical data is subject to high data protection. Anonymization is essential, but poses a challenge for some regions, such as the head, as identifying structures overlap with regions of clinical interest. Synthetic data offers a potential solution, but studies often lack rigorous evaluation of realism and utility. Therefore, we investigate to what extent synthetic data can replace real data in segmentation tasks. We employed head and neck cancer CT scans and brain glioma MRI scans from two large datasets. Synthetic data were generated using generative adversarial networks and diffusion models. We evaluated the quality of the synthetic data using MAE, MS-SSIM, Radiomics and a Visual Turing Test (VTT) performed by 5 radiologists and their usefulness in segmentation tasks using DSC. Radiomics indicates high fidelity of synthetic MRIs, but fall short in producing highly realistic CT tissue, with correlation coefficient of 0.8784 and 0.5461 for MRI and CT tumors, respectively. DSC results indicate limited utility of synthetic data: tumor segmentation achieved DSC=0.064 on CT and 0.834 on MRI, while bone segmentation a mean DSC=0.841. Relation between DSC and correlation is observed, but is limited by the complexity of the task. VTT results show synthetic CTs' utility, but with limited educational applications. Synthetic data can be used independently for the segmentation task, although limited by the complexity of the structures to segment. Advancing generative models to better tolerate heterogeneous inputs and learn subtle details is essential for enhancing their realism and expanding their application potential.
Efficient MedSAMs: Segment Anything in Medical Images on Laptop
Dec 20, 2024


Abstract:Promptable segmentation foundation models have emerged as a transformative approach to addressing the diverse needs in medical images, but most existing models require expensive computing, posing a big barrier to their adoption in clinical practice. In this work, we organized the first international competition dedicated to promptable medical image segmentation, featuring a large-scale dataset spanning nine common imaging modalities from over 20 different institutions. The top teams developed lightweight segmentation foundation models and implemented an efficient inference pipeline that substantially reduced computational requirements while maintaining state-of-the-art segmentation accuracy. Moreover, the post-challenge phase advanced the algorithms through the design of performance booster and reproducibility tasks, resulting in improved algorithms and validated reproducibility of the winning solution. Furthermore, the best-performing algorithms have been incorporated into the open-source software with a user-friendly interface to facilitate clinical adoption. The data and code are publicly available to foster the further development of medical image segmentation foundation models and pave the way for impactful real-world applications.
Cyto R-CNN and CytoNuke Dataset: Towards reliable whole-cell segmentation in bright-field histological images
Feb 04, 2024Abstract:Background: Cell segmentation in bright-field histological slides is a crucial topic in medical image analysis. Having access to accurate segmentation allows researchers to examine the relationship between cellular morphology and clinical observations. Unfortunately, most segmentation methods known today are limited to nuclei and cannot segmentate the cytoplasm. Material & Methods: We present a new network architecture Cyto R-CNN that is able to accurately segment whole cells (with both the nucleus and the cytoplasm) in bright-field images. We also present a new dataset CytoNuke, consisting of multiple thousand manual annotations of head and neck squamous cell carcinoma cells. Utilizing this dataset, we compared the performance of Cyto R-CNN to other popular cell segmentation algorithms, including QuPath's built-in algorithm, StarDist and Cellpose. To evaluate segmentation performance, we calculated AP50, AP75 and measured 17 morphological and staining-related features for all detected cells. We compared these measurements to the gold standard of manual segmentation using the Kolmogorov-Smirnov test. Results: Cyto R-CNN achieved an AP50 of 58.65% and an AP75 of 11.56% in whole-cell segmentation, outperforming all other methods (QuPath $19.46/0.91\%$; StarDist $45.33/2.32\%$; Cellpose $31.85/5.61\%$). Cell features derived from Cyto R-CNN showed the best agreement to the gold standard ($\bar{D} = 0.15$) outperforming QuPath ($\bar{D} = 0.22$), StarDist ($\bar{D} = 0.25$) and Cellpose ($\bar{D} = 0.23$). Conclusion: Our newly proposed Cyto R-CNN architecture outperforms current algorithms in whole-cell segmentation while providing more reliable cell measurements than any other model. This could improve digital pathology workflows, potentially leading to improved diagnosis. Moreover, our published dataset can be used to develop further models in the future.
Elastic Significant Bit Quantization and Acceleration for Deep Neural Networks
Sep 08, 2021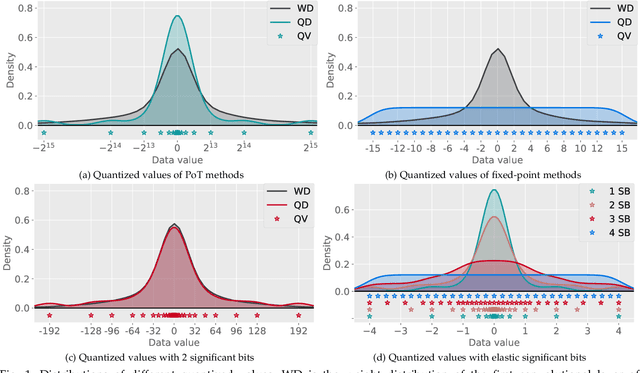
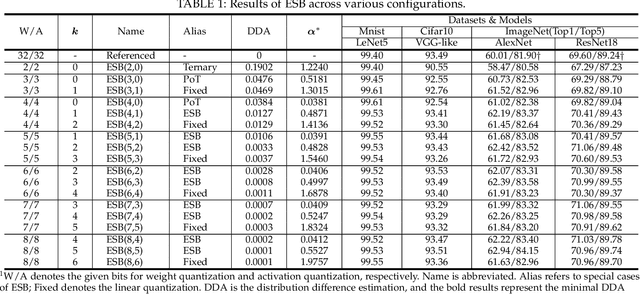
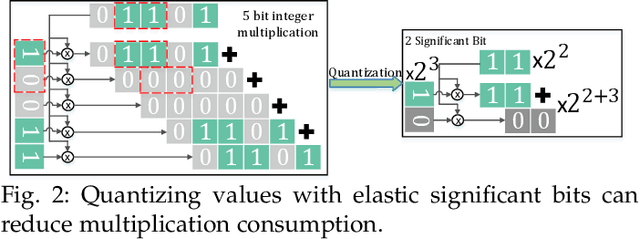
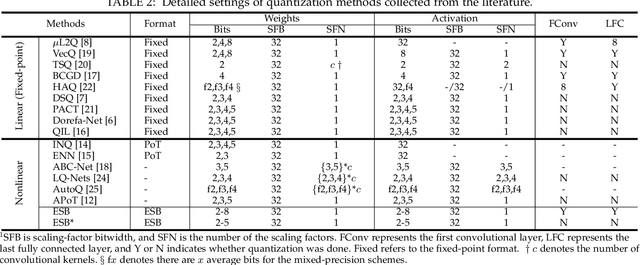
Abstract:Quantization has been proven to be a vital method for improving the inference efficiency of deep neural networks (DNNs). However, it is still challenging to strike a good balance between accuracy and efficiency while quantizing DNN weights or activation values from high-precision formats to their quantized counterparts. We propose a new method called elastic significant bit quantization (ESB) that controls the number of significant bits of quantized values to obtain better inference accuracy with fewer resources. We design a unified mathematical formula to constrain the quantized values of the ESB with a flexible number of significant bits. We also introduce a distribution difference aligner (DDA) to quantitatively align the distributions between the full-precision weight or activation values and quantized values. Consequently, ESB is suitable for various bell-shaped distributions of weights and activation of DNNs, thus maintaining a high inference accuracy. Benefitting from fewer significant bits of quantized values, ESB can reduce the multiplication complexity. We implement ESB as an accelerator and quantitatively evaluate its efficiency on FPGAs. Extensive experimental results illustrate that ESB quantization consistently outperforms state-of-the-art methods and achieves average accuracy improvements of 4.78%, 1.92%, and 3.56% over AlexNet, ResNet18, and MobileNetV2, respectively. Furthermore, ESB as an accelerator can achieve 10.95 GOPS peak performance of 1k LUTs without DSPs on the Xilinx ZCU102 FPGA platform. Compared with CPU, GPU, and state-of-the-art accelerators on FPGAs, the ESB accelerator can improve the energy efficiency by up to 65x, 11x, and 26x, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge