Donghang Lyu
CRUNet-MR-Univ: A Foundation Model for Diverse Cardiac MRI Reconstruction
Jan 07, 2026Abstract:In recent years, deep learning has attracted increasing at- tention in the field of Cardiac MRI (CMR) reconstruction due to its superior performance over traditional methods, particularly in handling higher acceleration factors, highlighting its potential for real-world clini- cal applications. However, current deep learning methods remain limited in generalizability. CMR scans exhibit wide variability in image contrast, sampling patterns, scanner vendors, anatomical structures, and disease types. Most existing models are designed to handle only a single or nar- row subset of these variations, leading to performance degradation when faced with distribution shifts. Therefore, it is beneficial to develop a unified model capable of generalizing across diverse CMR scenarios. To this end, we propose CRUNet-MR-Univ, a foundation model that lever- ages spatio-temporal correlations and prompt-based priors to effectively handle the full diversity of CMR scans. Our approach consistently out- performs baseline methods across a wide range of settings, highlighting its effectiveness and promise.
Towards Universal Learning-based Model for Cardiac Image Reconstruction: Summary of the CMRxRecon2024 Challenge
Mar 05, 2025Abstract:Cardiovascular magnetic resonance (CMR) offers diverse imaging contrasts for assessment of cardiac function and tissue characterization. However, acquiring each single CMR modality is often time-consuming, and comprehensive clinical protocols require multiple modalities with various sampling patterns, further extending the overall acquisition time and increasing susceptibility to motion artifacts. Existing deep learning-based reconstruction methods are often designed for specific acquisition parameters, which limits their ability to generalize across a variety of scan scenarios. As part of the CMRxRecon Series, the CMRxRecon2024 challenge provides diverse datasets encompassing multi-modality multi-view imaging with various sampling patterns, and a platform for the international community to develop and benchmark reconstruction solutions in two well-crafted tasks. Task 1 is a modality-universal setting, evaluating the out-of-distribution generalization of the reconstructed model, while Task 2 follows sampling-universal setting assessing the one-for-all adaptability of the universal model. Main contributions include providing the first and largest publicly available multi-modality, multi-view cardiac k-space dataset; developing a benchmarking platform that simulates clinical acceleration protocols, with a shared code library and tutorial for various k-t undersampling patterns and data processing; giving technical insights of enhanced data consistency based on physic-informed networks and adaptive prompt-learning embedding to be versatile to different clinical settings; additional finding on evaluation metrics to address the limitations of conventional ground-truth references in universal reconstruction tasks.
UPCMR: A Universal Prompt-guided Model for Random Sampling Cardiac MRI Reconstruction
Feb 18, 2025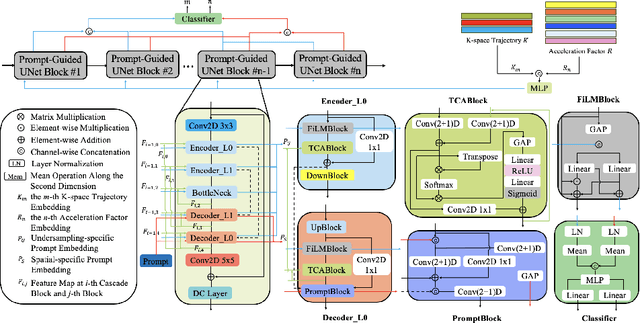
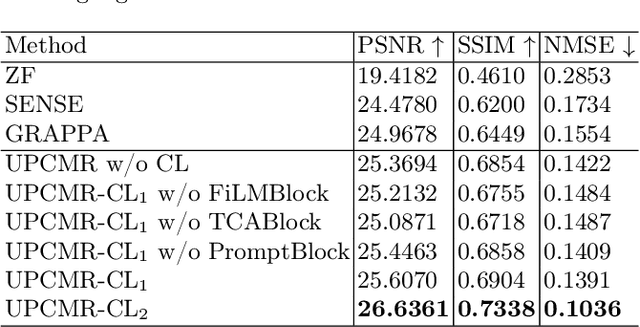
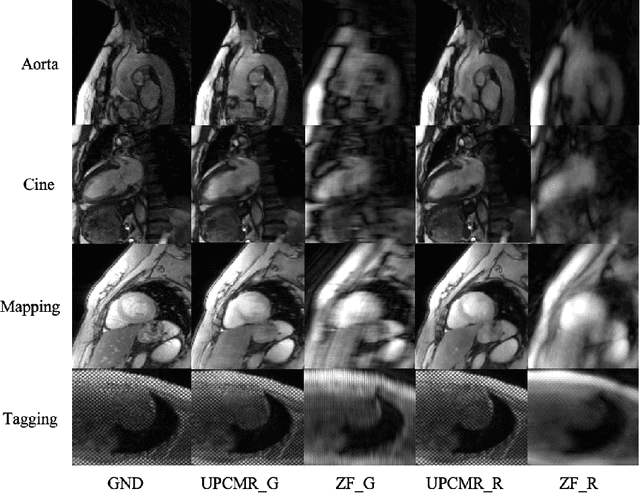
Abstract:Cardiac magnetic resonance imaging (CMR) is vital for diagnosing heart diseases, but long scan time remains a major drawback. To address this, accelerated imaging techniques have been introduced by undersampling k-space, which reduces the quality of the resulting images. Recent deep learning advancements aim to speed up scanning while preserving quality, but adapting to various sampling modes and undersampling factors remains challenging. Therefore, building a universal model is a promising direction. In this work, we introduce UPCMR, a universal unrolled model designed for CMR reconstruction. This model incorporates two kinds of learnable prompts, undersampling-specific prompt and spatial-specific prompt, and integrates them with a UNet structure in each block. Overall, by using the CMRxRecon2024 challenge dataset for training and validation, the UPCMR model highly enhances reconstructed image quality across all random sampling scenarios through an effective training strategy compared to some traditional methods, demonstrating strong adaptability potential for this task.
Efficient MedSAMs: Segment Anything in Medical Images on Laptop
Dec 20, 2024


Abstract:Promptable segmentation foundation models have emerged as a transformative approach to addressing the diverse needs in medical images, but most existing models require expensive computing, posing a big barrier to their adoption in clinical practice. In this work, we organized the first international competition dedicated to promptable medical image segmentation, featuring a large-scale dataset spanning nine common imaging modalities from over 20 different institutions. The top teams developed lightweight segmentation foundation models and implemented an efficient inference pipeline that substantially reduced computational requirements while maintaining state-of-the-art segmentation accuracy. Moreover, the post-challenge phase advanced the algorithms through the design of performance booster and reproducibility tasks, resulting in improved algorithms and validated reproducibility of the winning solution. Furthermore, the best-performing algorithms have been incorporated into the open-source software with a user-friendly interface to facilitate clinical adoption. The data and code are publicly available to foster the further development of medical image segmentation foundation models and pave the way for impactful real-world applications.
MCP-MedSAM: A Powerful Lightweight Medical Segment Anything Model Trained with a Single GPU in Just One Day
Dec 08, 2024



Abstract:Medical image segmentation involves partitioning medical images into meaningful regions, with a focus on identifying anatomical structures or abnormalities. It has broad applications in healthcare, and deep learning methods have enabled significant advancements in automating this process. Recently, the introduction of the Segmentation Anything Model (SAM), the first foundation model for segmentation task, has prompted researchers to adapt it for the medical domain to improve performance across various tasks. However, SAM's large model size and high GPU requirements hinder its scalability and development in the medical domain. To address these challenges, research has increasingly focused on lightweight adaptations of SAM to reduce its parameter count, enabling training with limited GPU resources while maintaining competitive segmentation performance. In this work, we propose MCP-MedSAM, a powerful and lightweight medical SAM model designed to be trainable on a single GPU within one day while delivering superior segmentation performance. Our method was trained and evaluated using a large-scale challenge dataset\footnote{\url{https://www.codabench.org/competitions/1847}\label{comp}}, compared to top-ranking methods on the challenge leaderboard, MCP-MedSAM achieved superior performance while requiring only one day of training on a single GPU. The code is publicly available at \url{https://github.com/dong845/MCP-MedSAM}.
Swin-LiteMedSAM: A Lightweight Box-Based Segment Anything Model for Large-Scale Medical Image Datasets
Sep 11, 2024Abstract:Medical imaging is essential for the diagnosis and treatment of diseases, with medical image segmentation as a subtask receiving high attention. However, automatic medical image segmentation models are typically task-specific and struggle to handle multiple scenarios, such as different imaging modalities and regions of interest. With the introduction of the Segment Anything Model (SAM), training a universal model for various clinical scenarios has become feasible. Recently, several Medical SAM (MedSAM) methods have been proposed, but these models often rely on heavy image encoders to achieve high performance, which may not be practical for real-world applications due to their high computational demands and slow inference speed. To address this issue, a lightweight version of the MedSAM (LiteMedSAM) can provide a viable solution, achieving high performance while requiring fewer resources and less time. In this work, we introduce Swin-LiteMedSAM, a new variant of LiteMedSAM. This model integrates the tiny Swin Transformer as the image encoder, incorporates multiple types of prompts, including box-based points and scribble generated from a given bounding box, and establishes skip connections between the image encoder and the mask decoder. In the \textit{Segment Anything in Medical Images on Laptop} challenge (CVPR 2024), our approach strikes a good balance between segmentation performance and speed, demonstrating significantly improved overall results across multiple modalities compared to the LiteMedSAM baseline provided by the challenge organizers. Our proposed model achieved a DSC score of \textbf{0.8678} and an NSD score of \textbf{0.8844} on the validation set. On the final test set, it attained a DSC score of \textbf{0.8193} and an NSD score of \textbf{0.8461}, securing fourth place in the challenge.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge