Nicola Pezzotti
UPCMR: A Universal Prompt-guided Model for Random Sampling Cardiac MRI Reconstruction
Feb 18, 2025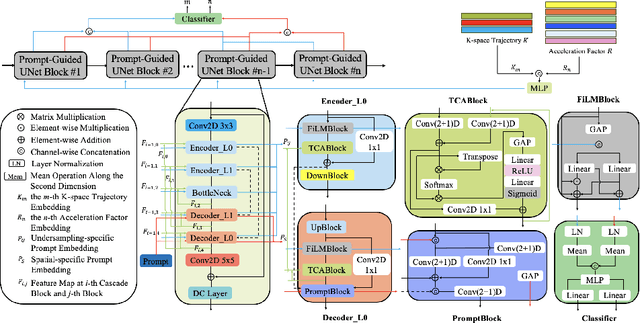
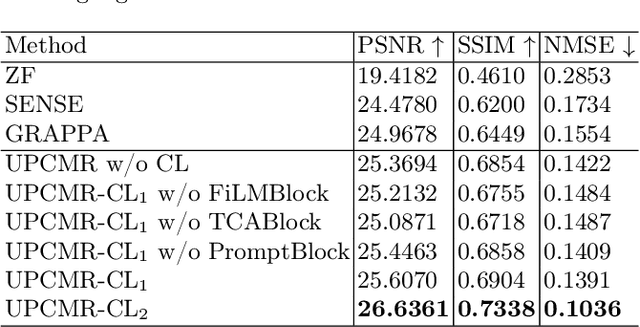
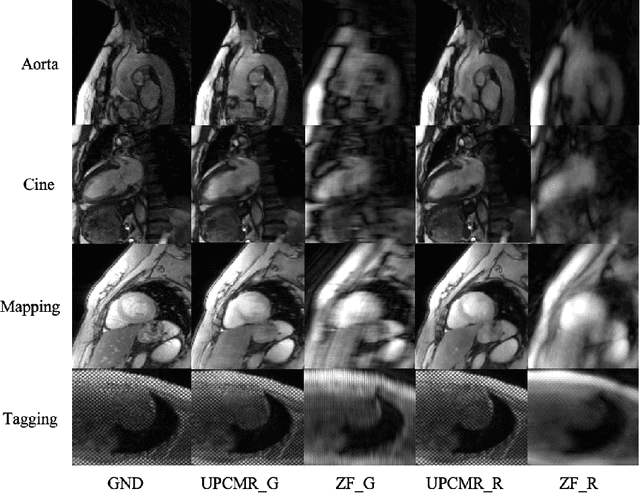
Abstract:Cardiac magnetic resonance imaging (CMR) is vital for diagnosing heart diseases, but long scan time remains a major drawback. To address this, accelerated imaging techniques have been introduced by undersampling k-space, which reduces the quality of the resulting images. Recent deep learning advancements aim to speed up scanning while preserving quality, but adapting to various sampling modes and undersampling factors remains challenging. Therefore, building a universal model is a promising direction. In this work, we introduce UPCMR, a universal unrolled model designed for CMR reconstruction. This model incorporates two kinds of learnable prompts, undersampling-specific prompt and spatial-specific prompt, and integrates them with a UNet structure in each block. Overall, by using the CMRxRecon2024 challenge dataset for training and validation, the UPCMR model highly enhances reconstructed image quality across all random sampling scenarios through an effective training strategy compared to some traditional methods, demonstrating strong adaptability potential for this task.
Progressive Monitoring of Generative Model Training Evolution
Dec 17, 2024Abstract:While deep generative models (DGMs) have gained popularity, their susceptibility to biases and other inefficiencies that lead to undesirable outcomes remains an issue. With their growing complexity, there is a critical need for early detection of issues to achieve desired results and optimize resources. Hence, we introduce a progressive analysis framework to monitor the training process of DGMs. Our method utilizes dimensionality reduction techniques to facilitate the inspection of latent representations, the generated and real distributions, and their evolution across training iterations. This monitoring allows us to pause and fix the training method if the representations or distributions progress undesirably. This approach allows for the analysis of a models' training dynamics and the timely identification of biases and failures, minimizing computational loads. We demonstrate how our method supports identifying and mitigating biases early in training a Generative Adversarial Network (GAN) and improving the quality of the generated data distribution.
NeRF-CA: Dynamic Reconstruction of X-ray Coronary Angiography with Extremely Sparse-views
Aug 29, 2024Abstract:Dynamic three-dimensional (4D) reconstruction from two-dimensional X-ray coronary angiography (CA) remains a significant clinical problem. Challenges include sparse-view settings, intra-scan motion, and complex vessel morphology such as structure sparsity and background occlusion. Existing CA reconstruction methods often require extensive user interaction or large training datasets. On the other hand, Neural Radiance Field (NeRF), a promising deep learning technique, has successfully reconstructed high-fidelity static scenes for natural and medical scenes. Recent work, however, identified that sparse-views, background occlusion, and dynamics still pose a challenge when applying NeRF in the X-ray angiography context. Meanwhile, many successful works for natural scenes propose regularization for sparse-view reconstruction or scene decomposition to handle dynamics. However, these techniques do not directly translate to the CA context, where both challenges and background occlusion are significant. This paper introduces NeRF-CA, the first step toward a 4D CA reconstruction method that achieves reconstructions from sparse coronary angiograms with cardiac motion. We leverage the motion of the coronary artery to decouple the scene into a dynamic coronary artery component and static background. We combine this scene decomposition with tailored regularization techniques. These techniques enforce the separation of the coronary artery from the background by enforcing dynamic structure sparsity and scene smoothness. By uniquely combining these approaches, we achieve 4D reconstructions from as few as four angiogram sequences. This setting aligns with clinical workflows while outperforming state-of-the-art X-ray sparse-view NeRF reconstruction techniques. We validate our approach quantitatively and qualitatively using 4D phantom datasets and ablation studies.
The Tree of Diffusion Life: Evolutionary Embeddings to Understand the Generation Process of Diffusion Models
Jun 25, 2024Abstract:Diffusion models generate high-quality samples by corrupting data with Gaussian noise and iteratively reconstructing it with deep learning, slowly transforming noisy images into refined outputs. Understanding this data evolution is important for interpretability but is complex due to its high-dimensional evolutionary nature. While traditional dimensionality reduction methods like t-distributed stochastic neighborhood embedding (t-SNE) aid in understanding high-dimensional spaces, they neglect evolutionary structure preservation. Hence, we propose Tree of Diffusion Life (TDL), a method to understand data evolution in the generative process of diffusion models. TDL samples a diffusion model's generative space via instances with varying prompts and employs image encoders to extract semantic meaning from these samples, projecting them to an intermediate space. It employs a novel evolutionary embedding algorithm that explicitly encodes the iterations while preserving the high-dimensional relations, facilitating the visualization of data evolution. This embedding leverages three metrics: a standard t-SNE loss to group semantically similar elements, a displacement loss to group elements from the same iteration step, and an instance alignment loss to align elements of the same instance across iterations. We present rectilinear and radial layouts to represent iterations, enabling comprehensive exploration. We assess various feature extractors and highlight TDL's potential with prominent diffusion models like GLIDE and Stable Diffusion with different prompt sets. TDL simplifies understanding data evolution within diffusion models, offering valuable insights into their functioning.
Unraveling the Temporal Dynamics of the Unet in Diffusion Models
Dec 17, 2023Abstract:Diffusion models have garnered significant attention since they can effectively learn complex multivariate Gaussian distributions, resulting in diverse, high-quality outcomes. They introduce Gaussian noise into training data and reconstruct the original data iteratively. Central to this iterative process is a single Unet, adapting across time steps to facilitate generation. Recent work revealed the presence of composition and denoising phases in this generation process, raising questions about the Unets' varying roles. Our study dives into the dynamic behavior of Unets within denoising diffusion probabilistic models (DDPM), focusing on (de)convolutional blocks and skip connections across time steps. We propose an analytical method to systematically assess the impact of time steps and core Unet components on the final output. This method eliminates components to study causal relations and investigate their influence on output changes. The main purpose is to understand the temporal dynamics and identify potential shortcuts during inference. Our findings provide valuable insights into the various generation phases during inference and shed light on the Unets' usage patterns across these phases. Leveraging these insights, we identify redundancies in GLIDE (an improved DDPM) and improve inference time by ~27% with minimal degradation in output quality. Our ultimate goal is to guide more informed optimization strategies for inference and influence new model designs.
Class-constrained t-SNE: Combining Data Features and Class Probabilities
Aug 26, 2023Abstract:Data features and class probabilities are two main perspectives when, e.g., evaluating model results and identifying problematic items. Class probabilities represent the likelihood that each instance belongs to a particular class, which can be produced by probabilistic classifiers or even human labeling with uncertainty. Since both perspectives are multi-dimensional data, dimensionality reduction (DR) techniques are commonly used to extract informative characteristics from them. However, existing methods either focus solely on the data feature perspective or rely on class probability estimates to guide the DR process. In contrast to previous work where separate views are linked to conduct the analysis, we propose a novel approach, class-constrained t-SNE, that combines data features and class probabilities in the same DR result. Specifically, we combine them by balancing two corresponding components in a cost function to optimize the positions of data points and iconic representation of classes -- class landmarks. Furthermore, an interactive user-adjustable parameter balances these two components so that users can focus on the weighted perspectives of interest and also empowers a smooth visual transition between varying perspectives to preserve the mental map. We illustrate its application potential in model evaluation and visual-interactive labeling. A comparative analysis is performed to evaluate the DR results.
Image Quality Assessment for Magnetic Resonance Imaging
Mar 15, 2022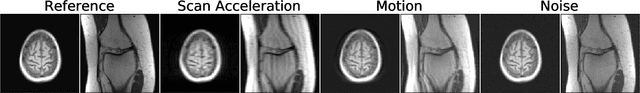
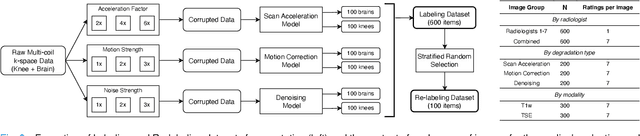
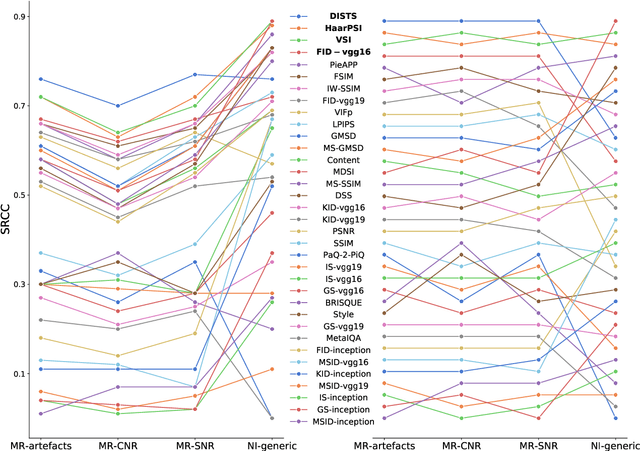
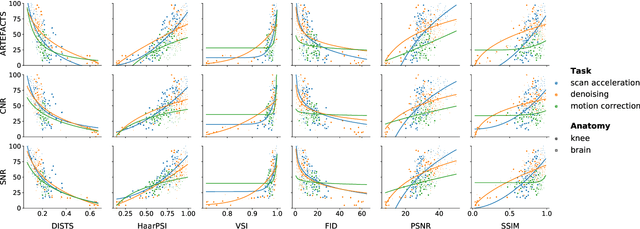
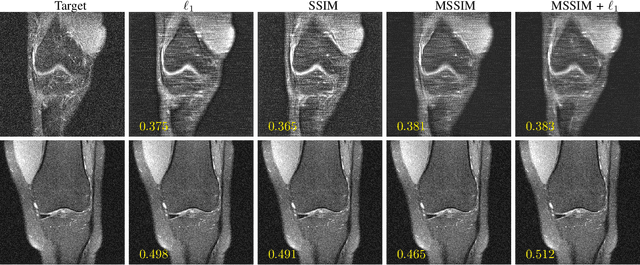
Abstract:Image quality assessment (IQA) algorithms aim to reproduce the human's perception of the image quality. The growing popularity of image enhancement, generation, and recovery models instigated the development of many methods to assess their performance. However, most IQA solutions are designed to predict image quality in the general domain, with the applicability to specific areas, such as medical imaging, remaining questionable. Moreover, the selection of these IQA metrics for a specific task typically involves intentionally induced distortions, such as manually added noise or artificial blurring; yet, the chosen metrics are then used to judge the output of real-life computer vision models. In this work, we aspire to fill these gaps by carrying out the most extensive IQA evaluation study for Magnetic Resonance Imaging (MRI) to date (14,700 subjective scores). We use outputs of neural network models trained to solve problems relevant to MRI, including image reconstruction in the scan acceleration, motion correction, and denoising. Seven trained radiologists assess these distorted images, with their verdicts then correlated with 35 different image quality metrics (full-reference, no-reference, and distribution-based metrics considered). Our emphasis is on reflecting the radiologist's perception of the reconstructed images, gauging the most diagnostically influential criteria for the quality of MRI scans: signal-to-noise ratio, contrast-to-noise ratio, and the presence of artifacts.
MimicBot: Combining Imitation and Reinforcement Learning to win in Bot Bowl
Aug 21, 2021
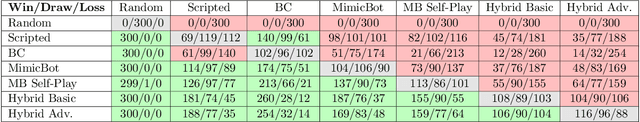
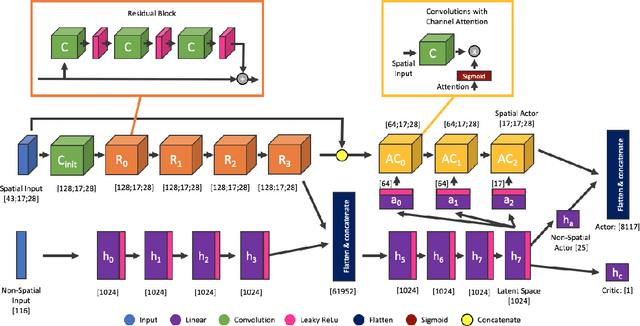
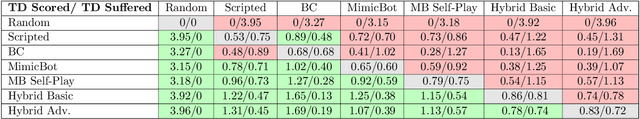
Abstract:This paper describe an hybrid agent trained to play in Fantasy Football AI which participated in the Bot Bowl III competition. The agent, MimicBot, is implemented using a specifically designed deep policy network and trained using a combination of imitation and reinforcement learning. Previous attempts in using a reinforcement learning approach in such context failed for a number of reasons, e.g. due to the intrinsic randomness in the environment and the large and uneven number of actions available, with a curriculum learning approach failing to consistently beat a randomly paying agent. Currently no machine learning approach can beat a scripted bot which makes use of the domain knowledge on the game. Our solution, thanks to an imitation learning and a hybrid decision-making process, consistently beat such scripted agents. Moreover we shed lights on how to more efficiently train in a reinforcement learning setting while drastically increasing sample efficiency. MimicBot is the winner of the Bot Bowl III competition, and it is currently the state-of-the-art solution.
An Adaptive Intelligence Algorithm for Undersampled Knee MRI Reconstruction: Application to the 2019 fastMRI Challenge
Apr 15, 2020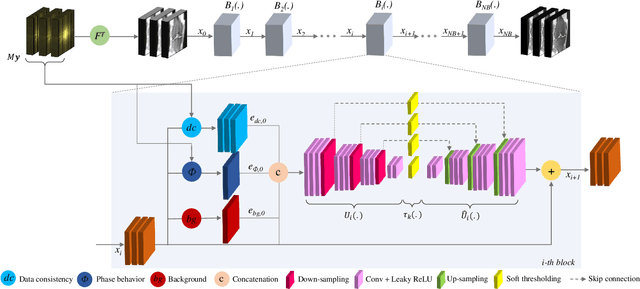
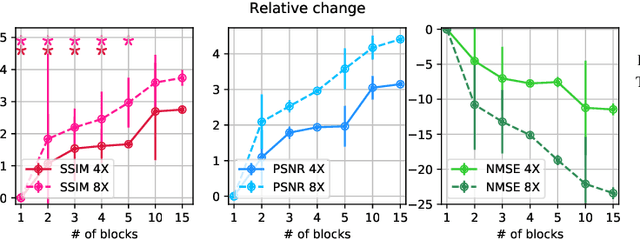

Abstract:Adaptive intelligence aims at empowering machine learning techniques with the additional use of domain knowledge. In this work, we present the application of adaptive intelligence to accelerate MR acquisition. Starting from undersampled k-space data, an iterative learning-based reconstruction scheme inspired by compressed sensing theory is used to reconstruct the images. We adopt deep neural networks to refine and correct prior reconstruction assumptions given the training data. The network was trained and tested on a knee MRI dataset from the 2019 fastMRI challenge organized by Facebook AI Research and NYU Langone Health. All submissions to the challenge were initially ranked based on similarity with a known groundtruth, after which the top 4 submissions were evaluated radiologically. Our method was evaluated by the fastMRI organizers on an independent challenge dataset. It ranked #1, shared #1, and #3 on respectively the 8x accelerated multi-coil, the 4x multi-coil, and the 4x single-coil track. This demonstrates the superior performance and wide applicability of the method.
Simple and Accurate Uncertainty Quantification from Bias-Variance Decomposition
Feb 13, 2020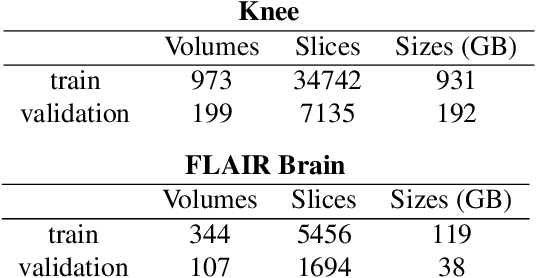
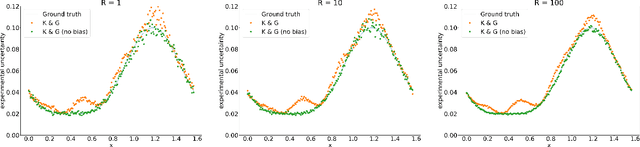
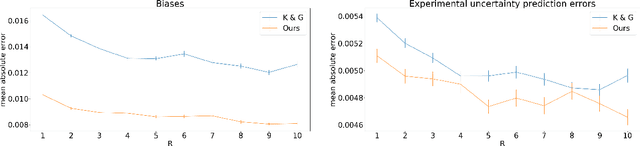
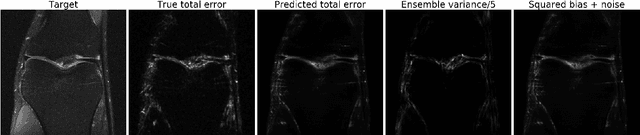
Abstract:Accurate uncertainty quantification is crucial for many applications where decisions are in play. Examples include medical diagnosis and self-driving vehicles. We propose a new method that is based directly on the bias-variance decomposition, where the parameter uncertainty is given by the variance of an ensemble divided by the number of members in the ensemble, and the aleatoric uncertainty plus the squared bias is estimated by training a separate model that is regressed directly on the errors of the predictor. We demonstrate that this simple sequential procedure provides much more accurate uncertainty estimates than the current state-of-the-art on two MRI reconstruction tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge