Danny Ruijters
OccluNet: Spatio-Temporal Deep Learning for Occlusion Detection on DSA
Aug 19, 2025Abstract:Accurate detection of vascular occlusions during endovascular thrombectomy (EVT) is critical in acute ischemic stroke (AIS). Interpretation of digital subtraction angiography (DSA) sequences poses challenges due to anatomical complexity and time constraints. This work proposes OccluNet, a spatio-temporal deep learning model that integrates YOLOX, a single-stage object detector, with transformer-based temporal attention mechanisms to automate occlusion detection in DSA sequences. We compared OccluNet with a YOLOv11 baseline trained on either individual DSA frames or minimum intensity projections. Two spatio-temporal variants were explored for OccluNet: pure temporal attention and divided space-time attention. Evaluation on DSA images from the MR CLEAN Registry revealed the model's capability to capture temporally consistent features, achieving precision and recall of 89.02% and 74.87%, respectively. OccluNet significantly outperformed the baseline models, and both attention variants attained similar performance. Source code is available at https://github.com/anushka-kore/OccluNet.git
ICA-based Resting-State Networks Obtained on Large Autism fMRI Dataset ABIDE
Dec 18, 2024Abstract:Functional magnetic resonance imaging (fMRI) has become instrumental in researching brain function. One application of fMRI is investigating potential neural features that distinguish people with autism spectrum disorder (ASD) from healthy controls. The Autism Brain Imaging Data Exchange (ABIDE) facilitates this research through its extensive data-sharing initiative. While ABIDE offers data preprocessed with various atlases, independent component analysis (ICA) for dimensionality reduction remains underutilized. We address this gap by presenting ICA-based resting-state networks (RSNs) from preprocessed scans from ABIDE, now publicly available: https://github.com/SjirSchielen/groupICAonABIDE. These RSNs unveil neural activation clusters without atlas constraints, offering a perspective on ASD analyses that complements the predominantly atlas-based literature. This contribution provides a valuable resource for further research into ASD, potentially aiding in developing new analytical approaches.
NeRF-CA: Dynamic Reconstruction of X-ray Coronary Angiography with Extremely Sparse-views
Aug 29, 2024Abstract:Dynamic three-dimensional (4D) reconstruction from two-dimensional X-ray coronary angiography (CA) remains a significant clinical problem. Challenges include sparse-view settings, intra-scan motion, and complex vessel morphology such as structure sparsity and background occlusion. Existing CA reconstruction methods often require extensive user interaction or large training datasets. On the other hand, Neural Radiance Field (NeRF), a promising deep learning technique, has successfully reconstructed high-fidelity static scenes for natural and medical scenes. Recent work, however, identified that sparse-views, background occlusion, and dynamics still pose a challenge when applying NeRF in the X-ray angiography context. Meanwhile, many successful works for natural scenes propose regularization for sparse-view reconstruction or scene decomposition to handle dynamics. However, these techniques do not directly translate to the CA context, where both challenges and background occlusion are significant. This paper introduces NeRF-CA, the first step toward a 4D CA reconstruction method that achieves reconstructions from sparse coronary angiograms with cardiac motion. We leverage the motion of the coronary artery to decouple the scene into a dynamic coronary artery component and static background. We combine this scene decomposition with tailored regularization techniques. These techniques enforce the separation of the coronary artery from the background by enforcing dynamic structure sparsity and scene smoothness. By uniquely combining these approaches, we achieve 4D reconstructions from as few as four angiogram sequences. This setting aligns with clinical workflows while outperforming state-of-the-art X-ray sparse-view NeRF reconstruction techniques. We validate our approach quantitatively and qualitatively using 4D phantom datasets and ablation studies.
AngioMoCo: Learning-based Motion Correction in Cerebral Digital Subtraction Angiography
Oct 09, 2023Abstract:Cerebral X-ray digital subtraction angiography (DSA) is the standard imaging technique for visualizing blood flow and guiding endovascular treatments. The quality of DSA is often negatively impacted by body motion during acquisition, leading to decreased diagnostic value. Time-consuming iterative methods address motion correction based on non-rigid registration, and employ sparse key points and non-rigidity penalties to limit vessel distortion. Recent methods alleviate subtraction artifacts by predicting the subtracted frame from the corresponding unsubtracted frame, but do not explicitly compensate for motion-induced misalignment between frames. This hinders the serial evaluation of blood flow, and often causes undesired vasculature and contrast flow alterations, leading to impeded usability in clinical practice. To address these limitations, we present AngioMoCo, a learning-based framework that generates motion-compensated DSA sequences from X-ray angiography. AngioMoCo integrates contrast extraction and motion correction, enabling differentiation between patient motion and intensity changes caused by contrast flow. This strategy improves registration quality while being substantially faster than iterative elastix-based methods. We demonstrate AngioMoCo on a large national multi-center dataset (MR CLEAN Registry) of clinically acquired angiographic images through comprehensive qualitative and quantitative analyses. AngioMoCo produces high-quality motion-compensated DSA, removing motion artifacts while preserving contrast flow. Code is publicly available at https://github.com/RuishengSu/AngioMoCo.
Spatio-Temporal U-Net for Cerebral Artery and Vein Segmentation in Digital Subtraction Angiography
Aug 03, 2022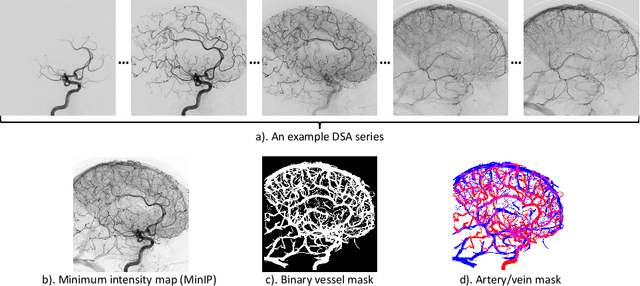
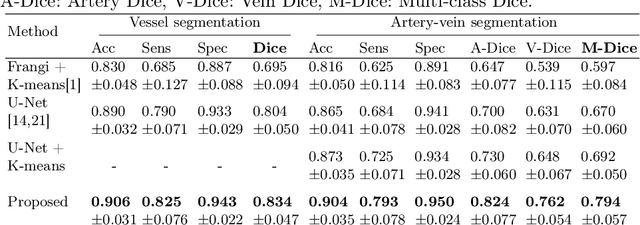
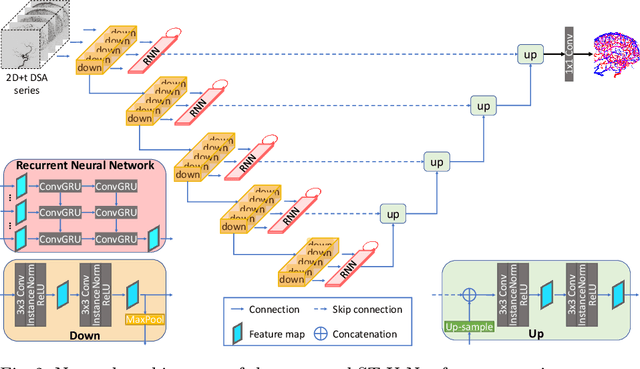
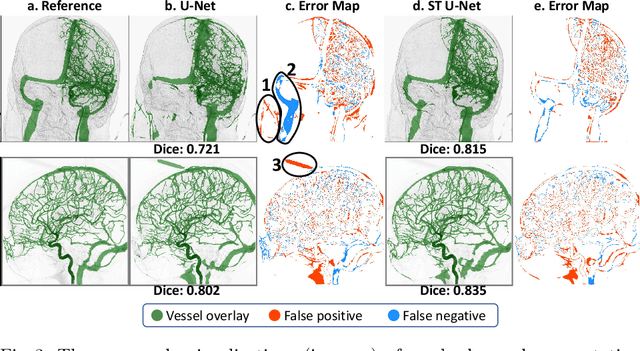
Abstract:X-ray digital subtraction angiography (DSA) is widely used for vessel and/or flow visualization and interventional guidance during endovascular treatment of patients with a stroke or aneurysm. To assist in peri-operative decision making as well as post-operative prognosis, automatic DSA analysis algorithms are being developed to obtain relevant image-based information. Such analyses include detection of vascular disease, evaluation of perfusion based on time intensity curves (TIC), and quantitative biomarker extraction for automated treatment evaluation in endovascular thrombectomy. Methodologically, such vessel-based analysis tasks may be facilitated by automatic and accurate artery-vein segmentation algorithms. The present work describes to the best of our knowledge the first study that addresses automatic artery-vein segmentation in DSA using deep learning. We propose a novel spatio-temporal U-Net (ST U-Net) architecture which integrates convolutional gated recurrent units (ConvGRU) in the contracting branch of U-Net. The network encodes a 2D+t DSA series of variable length and decodes it into a 2D segmentation image. On a multi-center routinely acquired dataset, the proposed method significantly outperformed U-Net (P<0.001) and traditional Frangi-based K-means clustering (P$<$0.001). Particularly in artery-vein segmentation, ST U-Net achieved a Dice coefficient of 0.794, surpassing the existing state-of-the-art methods by a margin of 12\%-20\%. Code will be made publicly available upon acceptance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge