Aad van der Lugt
Department of Radiology and Nuclear Medicine, Erasmus MC, Rotterdam, The Netherlands
An automated framework for brain vessel centerline extraction from CTA images
Jan 13, 2024Abstract:Accurate automated extraction of brain vessel centerlines from CTA images plays an important role in diagnosis and therapy of cerebrovascular diseases, such as stroke. However, this task remains challenging due to the complex cerebrovascular structure, the varying imaging quality, and vessel pathology effects. In this paper, we consider automatic lumen segmentation generation without additional annotation effort by physicians and more effective use of the generated lumen segmentation for improved centerline extraction performance. We propose an automated framework for brain vessel centerline extraction from CTA images. The framework consists of four major components: (1) pre-processing approaches that register CTA images with a CT atlas and divide these images into input patches, (2) lumen segmentation generation from annotated vessel centerlines using graph cuts and robust kernel regression, (3) a dual-branch topology-aware UNet (DTUNet) that can effectively utilize the annotated vessel centerlines and the generated lumen segmentation through a topology-aware loss (TAL) and its dual-branch design, and (4) post-processing approaches that skeletonize the predicted lumen segmentation. Extensive experiments on a multi-center dataset demonstrate that the proposed framework outperforms state-of-the-art methods in terms of average symmetric centerline distance (ASCD) and overlap (OV). Subgroup analyses further suggest that the proposed framework holds promise in clinical applications for stroke treatment. Code is publicly available at https://github.com/Liusj-gh/DTUNet.
AngioMoCo: Learning-based Motion Correction in Cerebral Digital Subtraction Angiography
Oct 09, 2023Abstract:Cerebral X-ray digital subtraction angiography (DSA) is the standard imaging technique for visualizing blood flow and guiding endovascular treatments. The quality of DSA is often negatively impacted by body motion during acquisition, leading to decreased diagnostic value. Time-consuming iterative methods address motion correction based on non-rigid registration, and employ sparse key points and non-rigidity penalties to limit vessel distortion. Recent methods alleviate subtraction artifacts by predicting the subtracted frame from the corresponding unsubtracted frame, but do not explicitly compensate for motion-induced misalignment between frames. This hinders the serial evaluation of blood flow, and often causes undesired vasculature and contrast flow alterations, leading to impeded usability in clinical practice. To address these limitations, we present AngioMoCo, a learning-based framework that generates motion-compensated DSA sequences from X-ray angiography. AngioMoCo integrates contrast extraction and motion correction, enabling differentiation between patient motion and intensity changes caused by contrast flow. This strategy improves registration quality while being substantially faster than iterative elastix-based methods. We demonstrate AngioMoCo on a large national multi-center dataset (MR CLEAN Registry) of clinically acquired angiographic images through comprehensive qualitative and quantitative analyses. AngioMoCo produces high-quality motion-compensated DSA, removing motion artifacts while preserving contrast flow. Code is publicly available at https://github.com/RuishengSu/AngioMoCo.
Spatio-Temporal U-Net for Cerebral Artery and Vein Segmentation in Digital Subtraction Angiography
Aug 03, 2022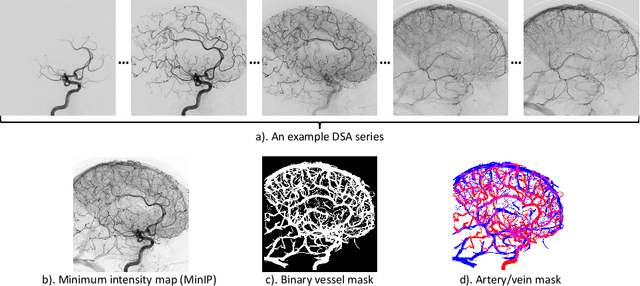
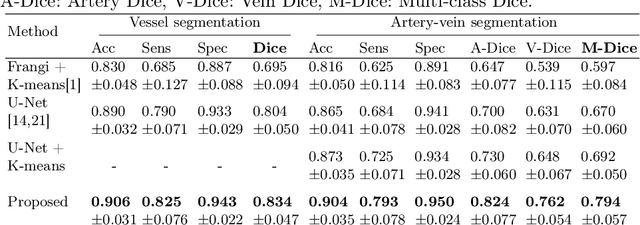
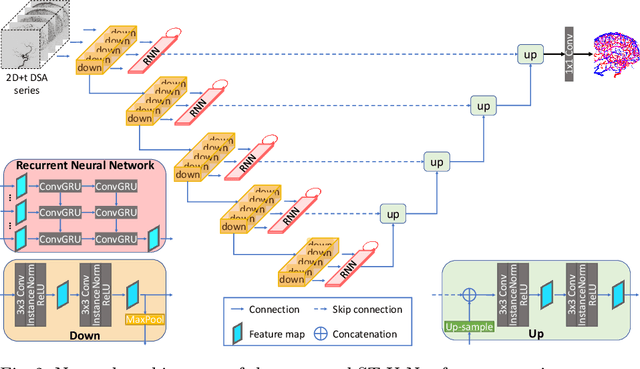
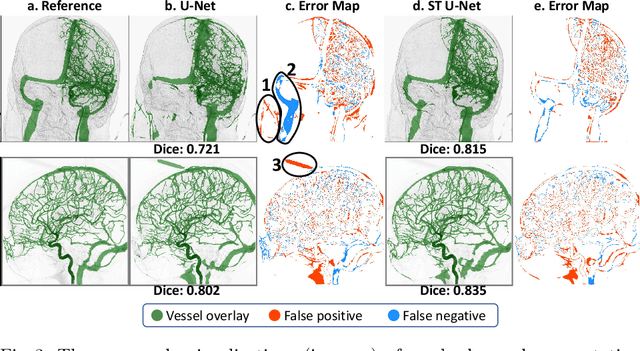
Abstract:X-ray digital subtraction angiography (DSA) is widely used for vessel and/or flow visualization and interventional guidance during endovascular treatment of patients with a stroke or aneurysm. To assist in peri-operative decision making as well as post-operative prognosis, automatic DSA analysis algorithms are being developed to obtain relevant image-based information. Such analyses include detection of vascular disease, evaluation of perfusion based on time intensity curves (TIC), and quantitative biomarker extraction for automated treatment evaluation in endovascular thrombectomy. Methodologically, such vessel-based analysis tasks may be facilitated by automatic and accurate artery-vein segmentation algorithms. The present work describes to the best of our knowledge the first study that addresses automatic artery-vein segmentation in DSA using deep learning. We propose a novel spatio-temporal U-Net (ST U-Net) architecture which integrates convolutional gated recurrent units (ConvGRU) in the contracting branch of U-Net. The network encodes a 2D+t DSA series of variable length and decodes it into a 2D segmentation image. On a multi-center routinely acquired dataset, the proposed method significantly outperformed U-Net (P<0.001) and traditional Frangi-based K-means clustering (P$<$0.001). Particularly in artery-vein segmentation, ST U-Net achieved a Dice coefficient of 0.794, surpassing the existing state-of-the-art methods by a margin of 12\%-20\%. Code will be made publicly available upon acceptance.
A Quantitative Comparison of Epistemic Uncertainty Maps Applied to Multi-Class Segmentation
Sep 22, 2021
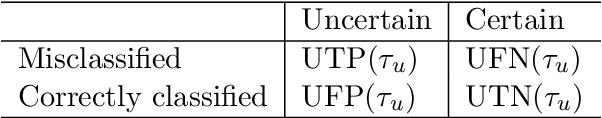
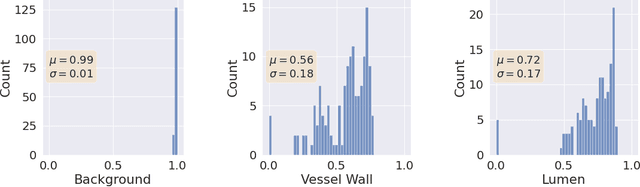
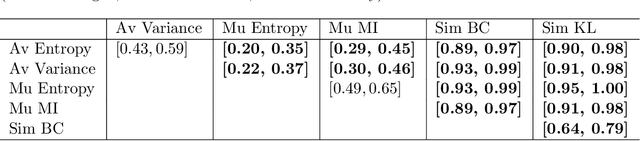
Abstract:Uncertainty assessment has gained rapid interest in medical image analysis. A popular technique to compute epistemic uncertainty is the Monte-Carlo (MC) dropout technique. From a network with MC dropout and a single input, multiple outputs can be sampled. Various methods can be used to obtain epistemic uncertainty maps from those multiple outputs. In the case of multi-class segmentation, the number of methods is even larger as epistemic uncertainty can be computed voxelwise per class or voxelwise per image. This paper highlights a systematic approach to define and quantitatively compare those methods in two different contexts: class-specific epistemic uncertainty maps (one value per image, voxel and class) and combined epistemic uncertainty maps (one value per image and voxel). We applied this quantitative analysis to a multi-class segmentation of the carotid artery lumen and vessel wall, on a multi-center, multi-scanner, multi-sequence dataset of (MR) images. We validated our analysis over 144 sets of hyperparameters of a model. Our main analysis considers the relationship between the order of the voxels sorted according to their epistemic uncertainty values and the misclassification of the prediction. Under this consideration, the comparison of combined uncertainty maps reveals that the multi-class entropy and the multi-class mutual information statistically out-perform the other combined uncertainty maps under study. In a class-specific scenario, the one-versus-all entropy statistically out-performs the class-wise entropy, the class-wise variance and the one versus all mutual information. The class-wise entropy statistically out-performs the other class-specific uncertainty maps in terms of calibration. We made a python package available to reproduce our analysis on different data and tasks.
Cross-Cohort Generalizability of Deep and Conventional Machine Learning for MRI-based Diagnosis and Prediction of Alzheimer's Disease
Dec 16, 2020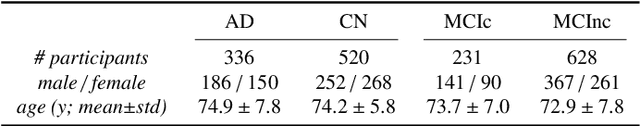
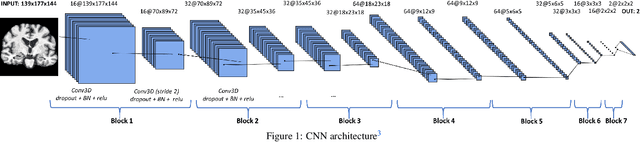
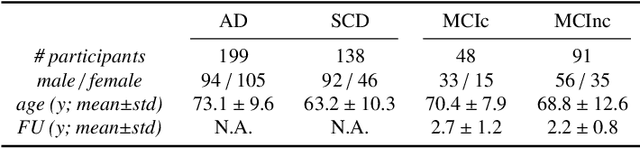
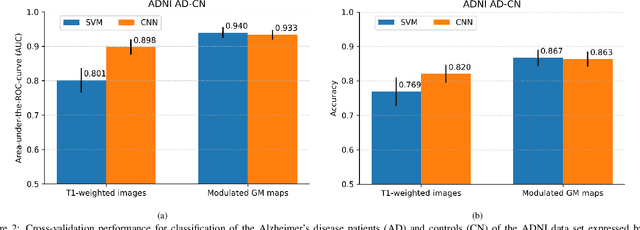
Abstract:This work validates the generalizability of MRI-based classification of Alzheimer's disease (AD) patients and controls (CN) to an external data set and to the task of prediction of conversion to AD in individuals with mild cognitive impairment (MCI). We used a conventional support vector machine (SVM) and a deep convolutional neural network (CNN) approach based on structural MRI scans that underwent either minimal pre-processing or more extensive pre-processing into modulated gray matter (GM) maps. Classifiers were optimized and evaluated using cross-validation in the ADNI (334 AD, 520 CN). Trained classifiers were subsequently applied to predict conversion to AD in ADNI MCI patients (231 converters, 628 non-converters) and in the independent Health-RI Parelsnoer data set. From this multi-center study representing a tertiary memory clinic population, we included 199 AD patients, 139 participants with subjective cognitive decline, 48 MCI patients converting to dementia, and 91 MCI patients who did not convert to dementia. AD-CN classification based on modulated GM maps resulted in a similar AUC for SVM (0.940) and CNN (0.933). Application to conversion prediction in MCI yielded significantly higher performance for SVM (0.756) than for CNN (0.742). In external validation, performance was slightly decreased. For AD-CN, it again gave similar AUCs for SVM (0.896) and CNN (0.876). For prediction in MCI, performances decreased for both SVM (0.665) and CNN (0.702). Both with SVM and CNN, classification based on modulated GM maps significantly outperformed classification based on minimally processed images. Deep and conventional classifiers performed equally well for AD classification and their performance decreased only slightly when applied to the external cohort. We expect that this work on external validation contributes towards translation of machine learning to clinical practice.
autoTICI: Automatic Brain Tissue Reperfusion Scoring on 2D DSA Images of Acute Ischemic Stroke Patients
Oct 06, 2020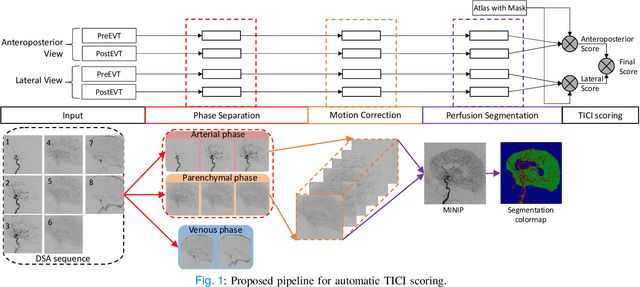
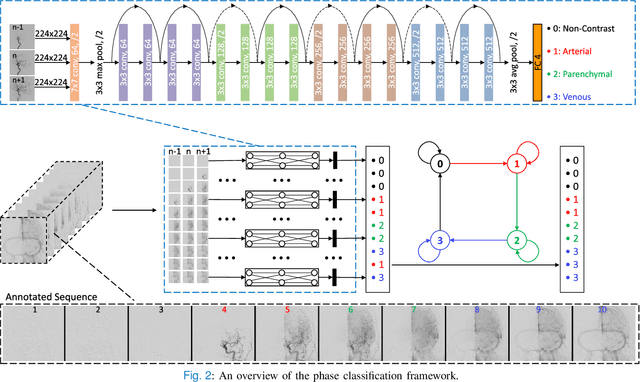
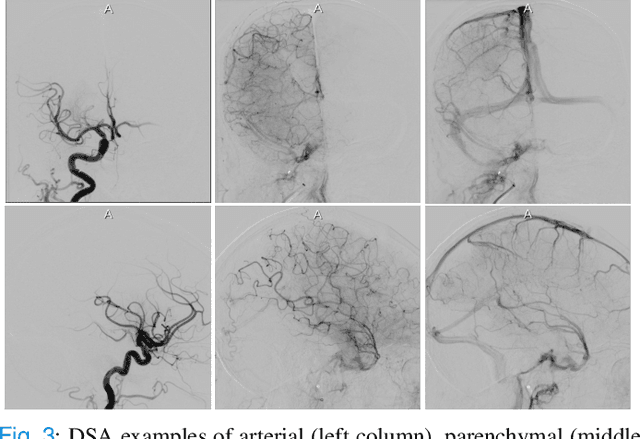
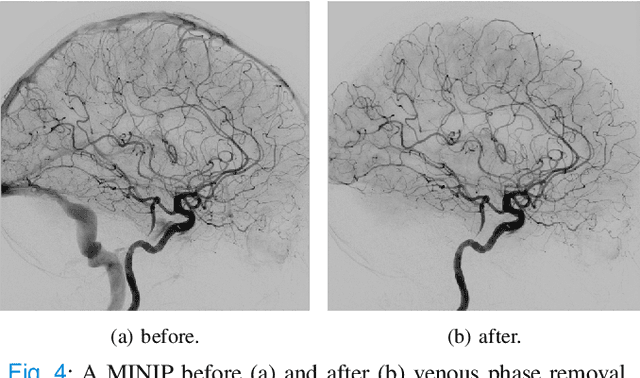
Abstract:The Thrombolysis in Cerebral Infarction (TICI) score is an important metric for reperfusion therapy assessment in acute ischemic stroke. It is commonly used as a technical outcome measure after endovascular treatment (EVT). Existing TICI scores are defined in coarse ordinal grades based on visual inspection, leading to inter- and intra-observer variation. In this work, we present autoTICI, an automatic and quantitative TICI scoring method. First, each digital subtraction angiography (DSA) sequence is separated into four phases (non-contrast, arterial, parenchymal and venous phase) using a multi-path convolutional neural network (CNN), which exploits spatio-temporal features. The network also incorporates sequence level label dependencies in the form of a state-transition matrix. Next, a minimum intensity map (MINIP) is computed using the motion corrected arterial and parenchymal frames. On the MINIP image, vessel, perfusion and background pixels are segmented. Finally, we quantify the autoTICI score as the ratio of reperfused pixels after EVT. On a routinely acquired multi-center dataset, the proposed autoTICI shows good correlation with the extended TICI (eTICI) reference with an average area under the curve (AUC) score of 0.81. The AUC score is 0.90 with respect to the dichotomized eTICI. In terms of clinical outcome prediction, we demonstrate that autoTICI is overall comparable to eTICI.
Prediction of final infarct volume from native CT perfusion and treatment parameters using deep learning
Dec 06, 2018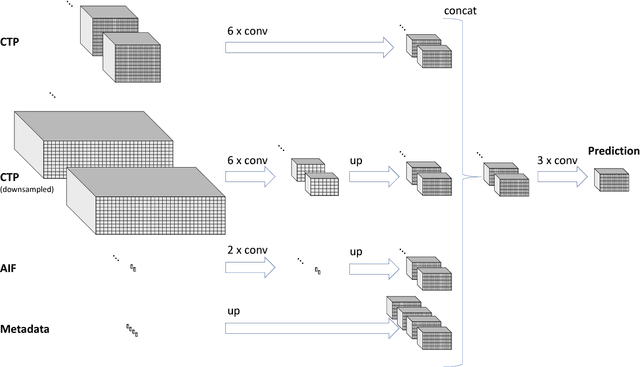
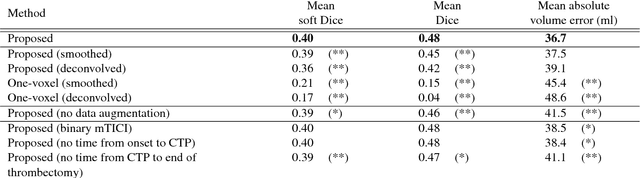
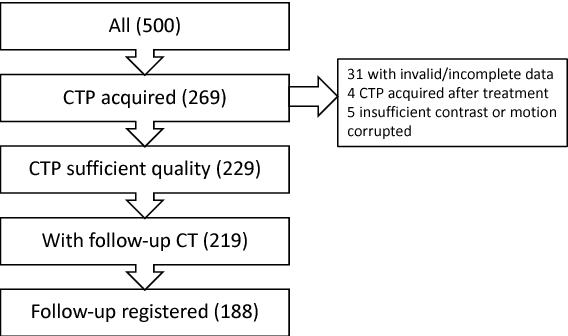
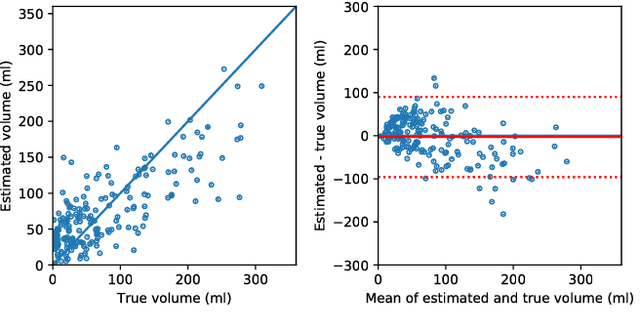
Abstract:CT Perfusion (CTP) imaging has gained importance in the diagnosis of acute stroke. Conventional perfusion analysis performs a deconvolution of the measurements and thresholds the perfusion parameters to determine the tissue status. We pursue a data-driven and deconvolution-free approach, where a deep neural network learns to predict the final infarct volume directly from the native CTP images and metadata such as the time parameters and treatment. This would allow clinicians to simulate various treatments and gain insight into predicted tissue status over time. We demonstrate on a multicenter dataset that our approach is able to predict the final infarct and effectively uses the metadata. An ablation study shows that using the native CTP measurements instead of the deconvolved measurements improves the prediction.
Segmentation of Intracranial Arterial Calcification with Deeply Supervised Residual Dropout Networks
Jun 04, 2017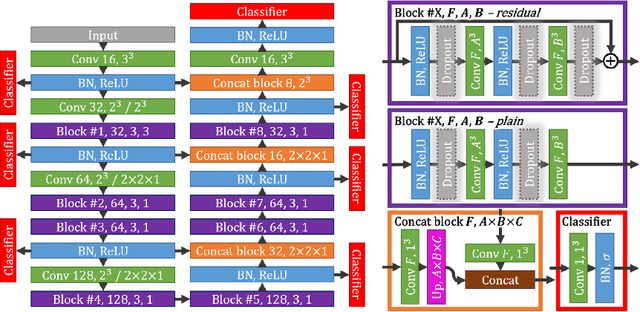
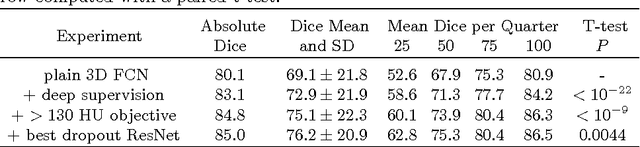
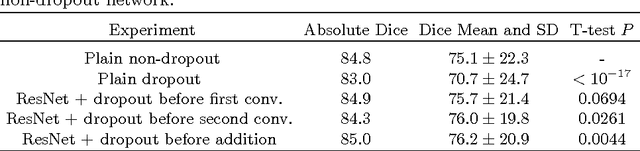
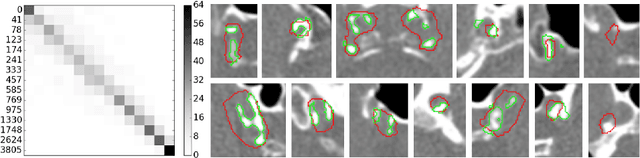
Abstract:Intracranial carotid artery calcification (ICAC) is a major risk factor for stroke, and might contribute to dementia and cognitive decline. Reliance on time-consuming manual annotation of ICAC hampers much demanded further research into the relationship between ICAC and neurological diseases. Automation of ICAC segmentation is therefore highly desirable, but difficult due to the proximity of the lesions to bony structures with a similar attenuation coefficient. In this paper, we propose a method for automatic segmentation of ICAC; the first to our knowledge. Our method is based on a 3D fully convolutional neural network that we extend with two regularization techniques. Firstly, we use deep supervision (hidden layers supervision) to encourage discriminative features in the hidden layers. Secondly, we augment the network with skip connections, as in the recently developed ResNet, and dropout layers, inserted in a way that skip connections circumvent them. We investigate the effect of skip connections and dropout. In addition, we propose a simple problem-specific modification of the network objective function that restricts the focus to the most important image regions and simplifies the optimization. We train and validate our model using 882 CT scans and test on 1,000. Our regularization techniques and objective improve the average Dice score by 7.1%, yielding an average Dice of 76.2% and 97.7% correlation between predicted ICAC volumes and manual annotations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge