Tingying Peng
Multimodal Whole Slide Foundation Model for Pathology
Nov 29, 2024



Abstract:The field of computational pathology has been transformed with recent advances in foundation models that encode histopathology region-of-interests (ROIs) into versatile and transferable feature representations via self-supervised learning (SSL). However, translating these advancements to address complex clinical challenges at the patient and slide level remains constrained by limited clinical data in disease-specific cohorts, especially for rare clinical conditions. We propose TITAN, a multimodal whole slide foundation model pretrained using 335,645 WSIs via visual self-supervised learning and vision-language alignment with corresponding pathology reports and 423,122 synthetic captions generated from a multimodal generative AI copilot for pathology. Without any finetuning or requiring clinical labels, TITAN can extract general-purpose slide representations and generate pathology reports that generalize to resource-limited clinical scenarios such as rare disease retrieval and cancer prognosis. We evaluate TITAN on diverse clinical tasks and find that TITAN outperforms both ROI and slide foundation models across machine learning settings such as linear probing, few-shot and zero-shot classification, rare cancer retrieval and cross-modal retrieval, and pathology report generation.
Fast Context-Based Low-Light Image Enhancement via Neural Implicit Representations
Jul 17, 2024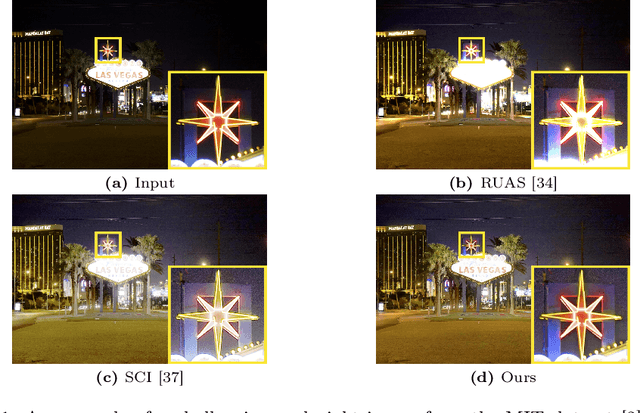
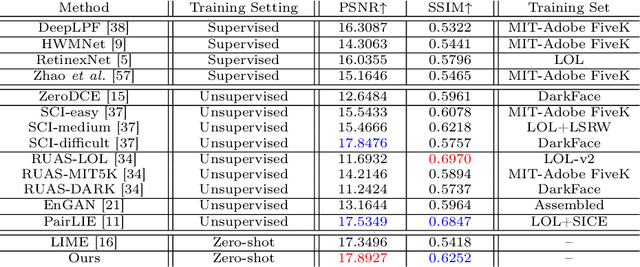
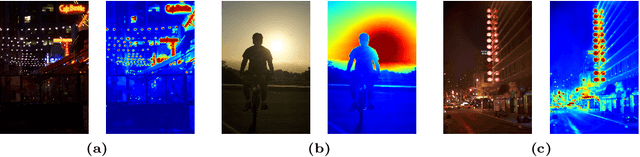
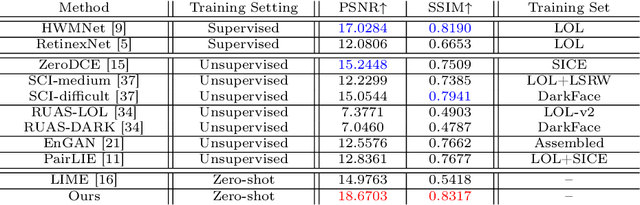
Abstract:Current deep learning-based low-light image enhancement methods often struggle with high-resolution images, and fail to meet the practical demands of visual perception across diverse and unseen scenarios. In this paper, we introduce a novel approach termed CoLIE, which redefines the enhancement process through mapping the 2D coordinates of an underexposed image to its illumination component, conditioned on local context. We propose a reconstruction of enhanced-light images within the HSV space utilizing an implicit neural function combined with an embedded guided filter, thereby significantly reducing computational overhead. Moreover, we introduce a single image-based training loss function to enhance the model's adaptability to various scenes, further enhancing its practical applicability. Through rigorous evaluations, we analyze the properties of our proposed framework, demonstrating its superiority in both image quality and scene adaptability. Furthermore, our evaluation extends to applications in downstream tasks within low-light scenarios, underscoring the practical utility of CoLIE. The source code is available at https://github.com/ctom2/colie.
DinoBloom: A Foundation Model for Generalizable Cell Embeddings in Hematology
Apr 07, 2024Abstract:In hematology, computational models offer significant potential to improve diagnostic accuracy, streamline workflows, and reduce the tedious work of analyzing single cells in peripheral blood or bone marrow smears. However, clinical adoption of computational models has been hampered by the lack of generalization due to large batch effects, small dataset sizes, and poor performance in transfer learning from natural images. To address these challenges, we introduce DinoBloom, the first foundation model for single cell images in hematology, utilizing a tailored DINOv2 pipeline. Our model is built upon an extensive collection of 13 diverse, publicly available datasets of peripheral blood and bone marrow smears, the most substantial open-source cohort in hematology so far, comprising over 380,000 white blood cell images. To assess its generalization capability, we evaluate it on an external dataset with a challenging domain shift. We show that our model outperforms existing medical and non-medical vision models in (i) linear probing and k-nearest neighbor evaluations for cell-type classification on blood and bone marrow smears and (ii) weakly supervised multiple instance learning for acute myeloid leukemia subtyping by a large margin. A family of four DinoBloom models (small, base, large, and giant) can be adapted for a wide range of downstream applications, be a strong baseline for classification problems, and facilitate the assessment of batch effects in new datasets. All models are available at github.com/marrlab/DinoBloom.
B-Cos Aligned Transformers Learn Human-Interpretable Features
Jan 18, 2024Abstract:Vision Transformers (ViTs) and Swin Transformers (Swin) are currently state-of-the-art in computational pathology. However, domain experts are still reluctant to use these models due to their lack of interpretability. This is not surprising, as critical decisions need to be transparent and understandable. The most common approach to understanding transformers is to visualize their attention. However, attention maps of ViTs are often fragmented, leading to unsatisfactory explanations. Here, we introduce a novel architecture called the B-cos Vision Transformer (BvT) that is designed to be more interpretable. It replaces all linear transformations with the B-cos transform to promote weight-input alignment. In a blinded study, medical experts clearly ranked BvTs above ViTs, suggesting that our network is better at capturing biomedically relevant structures. This is also true for the B-cos Swin Transformer (Bwin). Compared to the Swin Transformer, it even improves the F1-score by up to 4.7% on two public datasets.
Low-resource finetuning of foundation models beats state-of-the-art in histopathology
Jan 09, 2024Abstract:To handle the large scale of whole slide images in computational pathology, most approaches first tessellate the images into smaller patches, extract features from these patches, and finally aggregate the feature vectors with weakly-supervised learning. The performance of this workflow strongly depends on the quality of the extracted features. Recently, foundation models in computer vision showed that leveraging huge amounts of data through supervised or self-supervised learning improves feature quality and generalizability for a variety of tasks. In this study, we benchmark the most popular vision foundation models as feature extractors for histopathology data. We evaluate the models in two settings: slide-level classification and patch-level classification. We show that foundation models are a strong baseline. Our experiments demonstrate that by finetuning a foundation model on a single GPU for only two hours or three days depending on the dataset, we can match or outperform state-of-the-art feature extractors for computational pathology. These findings imply that even with little resources one can finetune a feature extractor tailored towards a specific downstream task and dataset. This is a considerable shift from the current state, where only few institutions with large amounts of resources and datasets are able to train a feature extractor. We publish all code used for training and evaluation as well as the finetuned models.
HiFi-Syn: Hierarchical Granularity Discrimination for High-Fidelity Synthesis of MR Images with Structure Preservation
Nov 21, 2023



Abstract:Synthesizing medical images while preserving their structural information is crucial in medical research. In such scenarios, the preservation of anatomical content becomes especially important. Although recent advances have been made by incorporating instance-level information to guide translation, these methods overlook the spatial coherence of structural-level representation and the anatomical invariance of content during translation. To address these issues, we introduce hierarchical granularity discrimination, which exploits various levels of semantic information present in medical images. Our strategy utilizes three levels of discrimination granularity: pixel-level discrimination using a Brain Memory Bank, structure-level discrimination on each brain structure with a re-weighting strategy to focus on hard samples, and global-level discrimination to ensure anatomical consistency during translation. The image translation performance of our strategy has been evaluated on three independent datasets (UK Biobank, IXI, and BraTS 2018), and it has outperformed state-of-the-art algorithms. Particularly, our model excels not only in synthesizing normal structures but also in handling abnormal (pathological) structures, such as brain tumors, despite the variations in contrast observed across different imaging modalities due to their pathological characteristics. The diagnostic value of synthesized MR images containing brain tumors has been evaluated by radiologists. This indicates that our model may offer an alternative solution in scenarios where specific MR modalities of patients are unavailable. Extensive experiments further demonstrate the versatility of our method, providing unique insights into medical image translation.
Leveraging Classic Deconvolution and Feature Extraction in Zero-Shot Image Restoration
Oct 03, 2023Abstract:Non-blind deconvolution aims to restore a sharp image from its blurred counterpart given an obtained kernel. Existing deep neural architectures are often built based on large datasets of sharp ground truth images and trained with supervision. Sharp, high quality ground truth images, however, are not always available, especially for biomedical applications. This severely hampers the applicability of current approaches in practice. In this paper, we propose a novel non-blind deconvolution method that leverages the power of deep learning and classic iterative deconvolution algorithms. Our approach combines a pre-trained network to extract deep features from the input image with iterative Richardson-Lucy deconvolution steps. Subsequently, a zero-shot optimisation process is employed to integrate the deconvolved features, resulting in a high-quality reconstructed image. By performing the preliminary reconstruction with the classic iterative deconvolution method, we can effectively utilise a smaller network to produce the final image, thus accelerating the reconstruction whilst reducing the demand for valuable computational resources. Our method demonstrates significant improvements in various real-world applications non-blind deconvolution tasks.
BigFUSE: Global Context-Aware Image Fusion in Dual-View Light-Sheet Fluorescence Microscopy with Image Formation Prior
Sep 05, 2023Abstract:Light-sheet fluorescence microscopy (LSFM), a planar illumination technique that enables high-resolution imaging of samples, experiences defocused image quality caused by light scattering when photons propagate through thick tissues. To circumvent this issue, dualview imaging is helpful. It allows various sections of the specimen to be scanned ideally by viewing the sample from opposing orientations. Recent image fusion approaches can then be applied to determine in-focus pixels by comparing image qualities of two views locally and thus yield spatially inconsistent focus measures due to their limited field-of-view. Here, we propose BigFUSE, a global context-aware image fuser that stabilizes image fusion in LSFM by considering the global impact of photon propagation in the specimen while determining focus-defocus based on local image qualities. Inspired by the image formation prior in dual-view LSFM, image fusion is considered as estimating a focus-defocus boundary using Bayes Theorem, where (i) the effect of light scattering onto focus measures is included within Likelihood; and (ii) the spatial consistency regarding focus-defocus is imposed in Prior. The expectation-maximum algorithm is then adopted to estimate the focus-defocus boundary. Competitive experimental results show that BigFUSE is the first dual-view LSFM fuser that is able to exclude structured artifacts when fusing information, highlighting its abilities of automatic image fusion.
LUCYD: A Feature-Driven Richardson-Lucy Deconvolution Network
Jul 16, 2023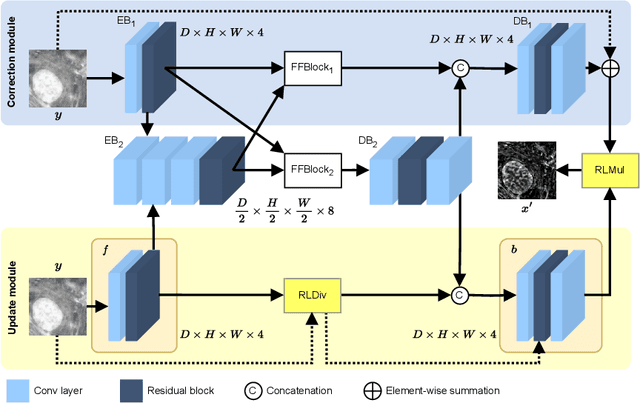
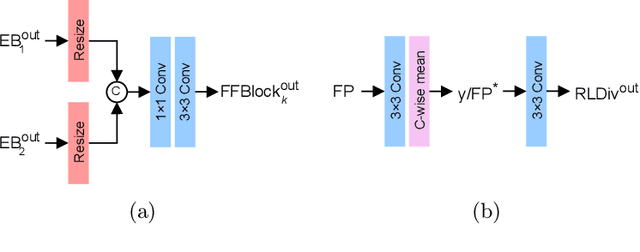
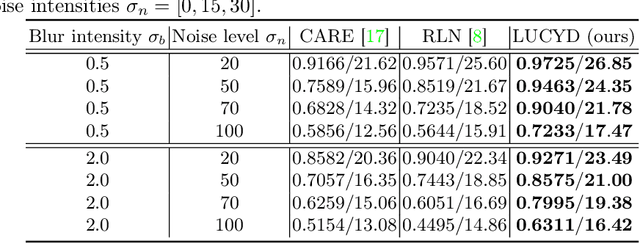
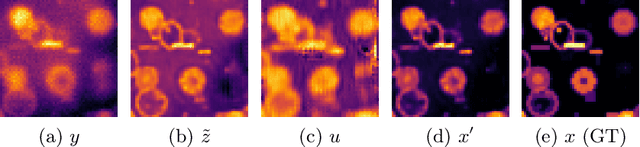
Abstract:The process of acquiring microscopic images in life sciences often results in image degradation and corruption, characterised by the presence of noise and blur, which poses significant challenges in accurately analysing and interpreting the obtained data. This paper proposes LUCYD, a novel method for the restoration of volumetric microscopy images that combines the Richardson-Lucy deconvolution formula and the fusion of deep features obtained by a fully convolutional network. By integrating the image formation process into a feature-driven restoration model, the proposed approach aims to enhance the quality of the restored images whilst reducing computational costs and maintaining a high degree of interpretability. Our results demonstrate that LUCYD outperforms the state-of-the-art methods in both synthetic and real microscopy images, achieving superior performance in terms of image quality and generalisability. We show that the model can handle various microscopy modalities and different imaging conditions by evaluating it on two different microscopy datasets, including volumetric widefield and light-sheet microscopy. Our experiments indicate that LUCYD can significantly improve resolution, contrast, and overall quality of microscopy images. Therefore, it can be a valuable tool for microscopy image restoration and can facilitate further research in various microscopy applications. We made the source code for the model accessible under https://github.com/ctom2/lucyd-deconvolution.
Training Transitive and Commutative Multimodal Transformers with LoReTTa
May 23, 2023Abstract:Collecting a multimodal dataset with two paired modalities A and B or B and C is difficult in practice. Obtaining a dataset with three aligned modalities A, B, and C is even more challenging. For example, some public medical datasets have only genetic sequences and microscopic images for one patient, and only genetic sequences and radiological images for another - but no dataset includes both microscopic and radiological images for the same patient. This makes it difficult to integrate and combine all modalities into a large pre-trained neural network. We introduce LoReTTa (Linking mOdalities with a tRansitive and commutativE pre-Training sTrAtegy) to address this understudied problem. Our self-supervised framework combines causal masked modeling with the rules of commutativity and transitivity to transition within and between different modalities. Thus, it can model the relation A -> C with A -> B -> C. Given a dataset containing only the disjoint combinations (A, B) and (B, C), we show that a transformer pre-trained with LoReTTa can handle any modality combination at inference time, including the never-seen pair (A, C) and the triplet (A, B, C). We evaluate our approach on a multimodal dataset derived from MNIST containing speech, vision, and language, as well as a real-world medical dataset containing mRNA, miRNA, and RPPA samples from TCGA. Compared to traditional pre-training methods, we observe up to a 100-point reduction in perplexity for autoregressive generation tasks and up to a 15% improvement in classification accuracy for previously unseen modality pairs during the pre-training phase.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge