Jan Huisken
Open Problem: Active Representation Learning
Jun 06, 2024
Abstract:In this work, we introduce the concept of Active Representation Learning, a novel class of problems that intertwines exploration and representation learning within partially observable environments. We extend ideas from Active Simultaneous Localization and Mapping (active SLAM), and translate them to scientific discovery problems, exemplified by adaptive microscopy. We explore the need for a framework that derives exploration skills from representations that are in some sense actionable, aiming to enhance the efficiency and effectiveness of data collection and model building in the natural sciences.
Leveraging Classic Deconvolution and Feature Extraction in Zero-Shot Image Restoration
Oct 03, 2023Abstract:Non-blind deconvolution aims to restore a sharp image from its blurred counterpart given an obtained kernel. Existing deep neural architectures are often built based on large datasets of sharp ground truth images and trained with supervision. Sharp, high quality ground truth images, however, are not always available, especially for biomedical applications. This severely hampers the applicability of current approaches in practice. In this paper, we propose a novel non-blind deconvolution method that leverages the power of deep learning and classic iterative deconvolution algorithms. Our approach combines a pre-trained network to extract deep features from the input image with iterative Richardson-Lucy deconvolution steps. Subsequently, a zero-shot optimisation process is employed to integrate the deconvolved features, resulting in a high-quality reconstructed image. By performing the preliminary reconstruction with the classic iterative deconvolution method, we can effectively utilise a smaller network to produce the final image, thus accelerating the reconstruction whilst reducing the demand for valuable computational resources. Our method demonstrates significant improvements in various real-world applications non-blind deconvolution tasks.
BigFUSE: Global Context-Aware Image Fusion in Dual-View Light-Sheet Fluorescence Microscopy with Image Formation Prior
Sep 05, 2023Abstract:Light-sheet fluorescence microscopy (LSFM), a planar illumination technique that enables high-resolution imaging of samples, experiences defocused image quality caused by light scattering when photons propagate through thick tissues. To circumvent this issue, dualview imaging is helpful. It allows various sections of the specimen to be scanned ideally by viewing the sample from opposing orientations. Recent image fusion approaches can then be applied to determine in-focus pixels by comparing image qualities of two views locally and thus yield spatially inconsistent focus measures due to their limited field-of-view. Here, we propose BigFUSE, a global context-aware image fuser that stabilizes image fusion in LSFM by considering the global impact of photon propagation in the specimen while determining focus-defocus based on local image qualities. Inspired by the image formation prior in dual-view LSFM, image fusion is considered as estimating a focus-defocus boundary using Bayes Theorem, where (i) the effect of light scattering onto focus measures is included within Likelihood; and (ii) the spatial consistency regarding focus-defocus is imposed in Prior. The expectation-maximum algorithm is then adopted to estimate the focus-defocus boundary. Competitive experimental results show that BigFUSE is the first dual-view LSFM fuser that is able to exclude structured artifacts when fusing information, highlighting its abilities of automatic image fusion.
LUCYD: A Feature-Driven Richardson-Lucy Deconvolution Network
Jul 16, 2023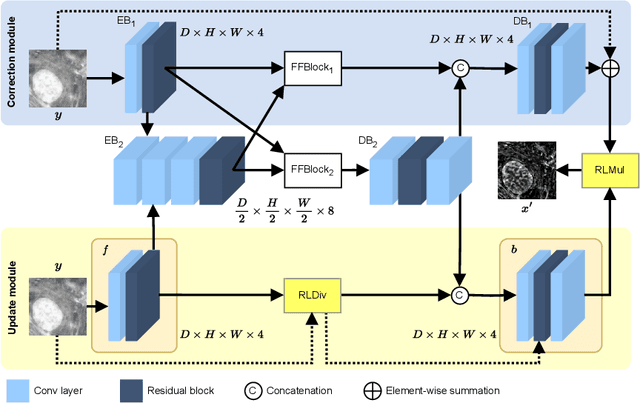
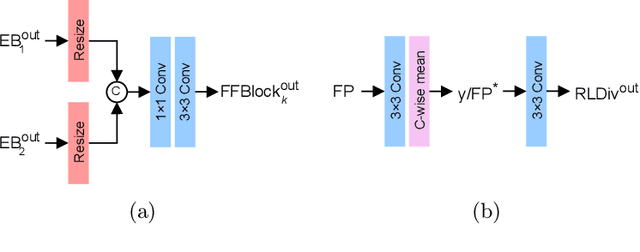
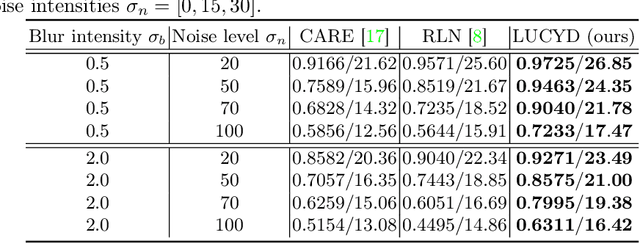
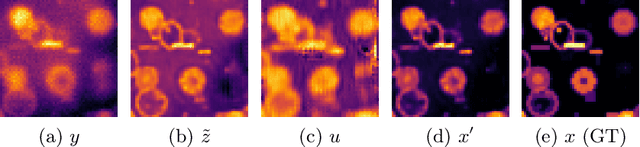
Abstract:The process of acquiring microscopic images in life sciences often results in image degradation and corruption, characterised by the presence of noise and blur, which poses significant challenges in accurately analysing and interpreting the obtained data. This paper proposes LUCYD, a novel method for the restoration of volumetric microscopy images that combines the Richardson-Lucy deconvolution formula and the fusion of deep features obtained by a fully convolutional network. By integrating the image formation process into a feature-driven restoration model, the proposed approach aims to enhance the quality of the restored images whilst reducing computational costs and maintaining a high degree of interpretability. Our results demonstrate that LUCYD outperforms the state-of-the-art methods in both synthetic and real microscopy images, achieving superior performance in terms of image quality and generalisability. We show that the model can handle various microscopy modalities and different imaging conditions by evaluating it on two different microscopy datasets, including volumetric widefield and light-sheet microscopy. Our experiments indicate that LUCYD can significantly improve resolution, contrast, and overall quality of microscopy images. Therefore, it can be a valuable tool for microscopy image restoration and can facilitate further research in various microscopy applications. We made the source code for the model accessible under https://github.com/ctom2/lucyd-deconvolution.
DeStripe: A Self2Self Spatio-Spectral Graph Neural Network with Unfolded Hessian for Stripe Artifact Removal in Light-sheet Microscopy
Jun 27, 2022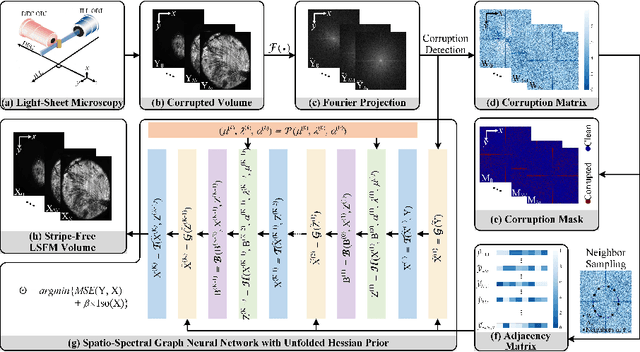
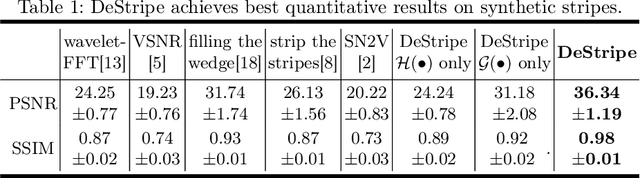
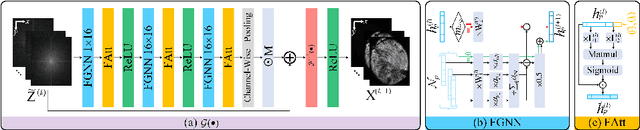
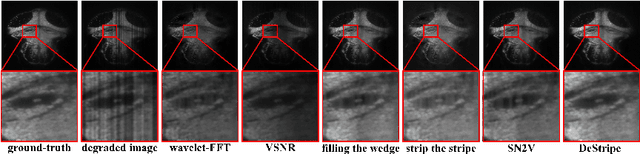
Abstract:Light-sheet fluorescence microscopy (LSFM) is a cutting-edge volumetric imaging technique that allows for three-dimensional imaging of mesoscopic samples with decoupled illumination and detection paths. Although the selective excitation scheme of such a microscope provides intrinsic optical sectioning that minimizes out-of-focus fluorescence background and sample photodamage, it is prone to light absorption and scattering effects, which results in uneven illumination and striping artifacts in the images adversely. To tackle this issue, in this paper, we propose a blind stripe artifact removal algorithm in LSFM, called DeStripe, which combines a self-supervised spatio-spectral graph neural network with unfolded Hessian prior. Specifically, inspired by the desirable properties of Fourier transform in condensing striping information into isolated values in the frequency domain, DeStripe firstly localizes the potentially corrupted Fourier coefficients by exploiting the structural difference between unidirectional stripe artifacts and more isotropic foreground images. Affected Fourier coefficients can then be fed into a graph neural network for recovery, with a Hessian regularization unrolled to further ensure structures in the standard image space are well preserved. Since in realistic, stripe-free LSFM barely exists with a standard image acquisition protocol, DeStripe is equipped with a Self2Self denoising loss term, enabling artifact elimination without access to stripe-free ground truth images. Competitive experimental results demonstrate the efficacy of DeStripe in recovering corrupted biomarkers in LSFM with both synthetic and real stripe artifacts.
Real-time multi-view deconvolution
Mar 27, 2015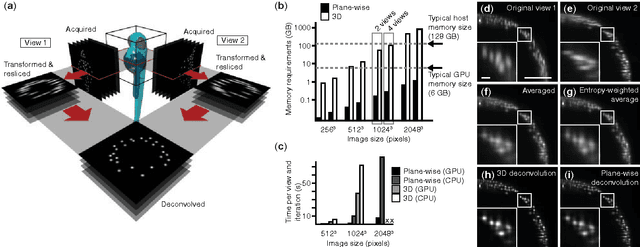
Abstract:In light-sheet microscopy, overall image content and resolution are improved by acquiring and fusing multiple views of the sample from different directions. State-of-the-art multi-view (MV) deconvolution employs the point spread functions (PSF) of the different views to simultaneously fuse and deconvolve the images in 3D, but processing takes a multiple of the acquisition time and constitutes the bottleneck in the imaging pipeline. Here we show that MV deconvolution in 3D can finally be achieved in real-time by reslicing the acquired data and processing cross-sectional planes individually on the massively parallel architecture of a graphics processing unit (GPU).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge