Nassir Navab
Computer Aided Medical Procedures, Technische Universit Munchen, Germany, Johns Hopkins University, Baltimore MD, USA
UniSurg: A Video-Native Foundation Model for Universal Understanding of Surgical Videos
Feb 05, 2026Abstract:While foundation models have advanced surgical video analysis, current approaches rely predominantly on pixel-level reconstruction objectives that waste model capacity on low-level visual details - such as smoke, specular reflections, and fluid motion - rather than semantic structures essential for surgical understanding. We present UniSurg, a video-native foundation model that shifts the learning paradigm from pixel-level reconstruction to latent motion prediction. Built on the Video Joint Embedding Predictive Architecture (V-JEPA), UniSurg introduces three key technical innovations tailored to surgical videos: 1) motion-guided latent prediction to prioritize semantically meaningful regions, 2) spatiotemporal affinity self-distillation to enforce relational consistency, and 3) feature diversity regularization to prevent representation collapse in texture-sparse surgical scenes. To enable large-scale pretraining, we curate UniSurg-15M, the largest surgical video dataset to date, comprising 3,658 hours of video from 50 sources across 13 anatomical regions. Extensive experiments across 17 benchmarks demonstrate that UniSurg significantly outperforms state-of-the-art methods on surgical workflow recognition (+14.6% F1 on EgoSurgery, +10.3% on PitVis), action triplet recognition (39.54% mAP-IVT on CholecT50), skill assessment, polyp segmentation, and depth estimation. These results establish UniSurg as a new standard for universal, motion-oriented surgical video understanding.
Where It Moves, It Matters: Referring Surgical Instrument Segmentation via Motion
Jan 18, 2026Abstract:Enabling intuitive, language-driven interaction with surgical scenes is a critical step toward intelligent operating rooms and autonomous surgical robotic assistance. However, the task of referring segmentation, localizing surgical instruments based on natural language descriptions, remains underexplored in surgical videos, with existing approaches struggling to generalize due to reliance on static visual cues and predefined instrument names. In this work, we introduce SurgRef, a novel motion-guided framework that grounds free-form language expressions in instrument motion, capturing how tools move and interact across time, rather than what they look like. This allows models to understand and segment instruments even under occlusion, ambiguity, or unfamiliar terminology. To train and evaluate SurgRef, we present Ref-IMotion, a diverse, multi-institutional video dataset with dense spatiotemporal masks and rich motion-centric expressions. SurgRef achieves state-of-the-art accuracy and generalization across surgical procedures, setting a new benchmark for robust, language-driven surgical video segmentation.
SurgGoal: Rethinking Surgical Planning Evaluation via Goal-Satisfiability
Jan 15, 2026Abstract:Surgical planning integrates visual perception, long-horizon reasoning, and procedural knowledge, yet it remains unclear whether current evaluation protocols reliably assess vision-language models (VLMs) in safety-critical settings. Motivated by a goal-oriented view of surgical planning, we define planning correctness via phase-goal satisfiability, where plan validity is determined by expert-defined surgical rules. Based on this definition, we introduce a multicentric meta-evaluation benchmark with valid procedural variations and invalid plans containing order and content errors. Using this benchmark, we show that sequence similarity metrics systematically misjudge planning quality, penalizing valid plans while failing to identify invalid ones. We therefore adopt a rule-based goal-satisfiability metric as a high-precision meta-evaluation reference to assess Video-LLMs under progressively constrained settings, revealing failures due to perception errors and under-constrained reasoning. Structural knowledge consistently improves performance, whereas semantic guidance alone is unreliable and benefits larger models only when combined with structural constraints.
COMPOSE: Hypergraph Cover Optimization for Multi-view 3D Human Pose Estimation
Jan 14, 2026Abstract:3D pose estimation from sparse multi-views is a critical task for numerous applications, including action recognition, sports analysis, and human-robot interaction. Optimization-based methods typically follow a two-stage pipeline, first detecting 2D keypoints in each view and then associating these detections across views to triangulate the 3D pose. Existing methods rely on mere pairwise associations to model this correspondence problem, treating global consistency between views (i.e., cycle consistency) as a soft constraint. Yet, reconciling these constraints for multiple views becomes brittle when spurious associations propagate errors. We thus propose COMPOSE, a novel framework that formulates multi-view pose correspondence matching as a hypergraph partitioning problem rather than through pairwise association. While the complexity of the resulting integer linear program grows exponentially in theory, we introduce an efficient geometric pruning strategy to substantially reduce the search space. COMPOSE achieves improvements of up to 23% in average precision over previous optimization-based methods and up to 11% over self-supervised end-to-end learned methods, offering a promising solution to a widely studied problem.
Visual Autoregressive Modelling for Monocular Depth Estimation
Dec 27, 2025Abstract:We propose a monocular depth estimation method based on visual autoregressive (VAR) priors, offering an alternative to diffusion-based approaches. Our method adapts a large-scale text-to-image VAR model and introduces a scale-wise conditional upsampling mechanism with classifier-free guidance. Our approach performs inference in ten fixed autoregressive stages, requiring only 74K synthetic samples for fine-tuning, and achieves competitive results. We report state-of-the-art performance in indoor benchmarks under constrained training conditions, and strong performance when applied to outdoor datasets. This work establishes autoregressive priors as a complementary family of geometry-aware generative models for depth estimation, highlighting advantages in data scalability, and adaptability to 3D vision tasks. Code available at "https://github.com/AmirMaEl/VAR-Depth".
ProtoFlow: Interpretable and Robust Surgical Workflow Modeling with Learned Dynamic Scene Graph Prototypes
Dec 16, 2025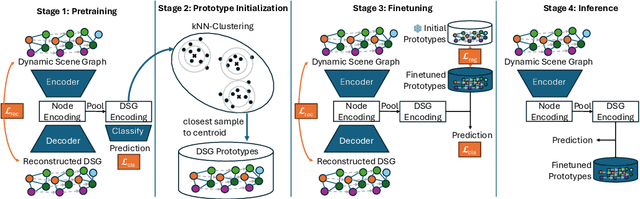
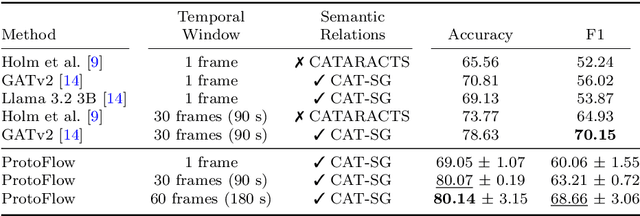
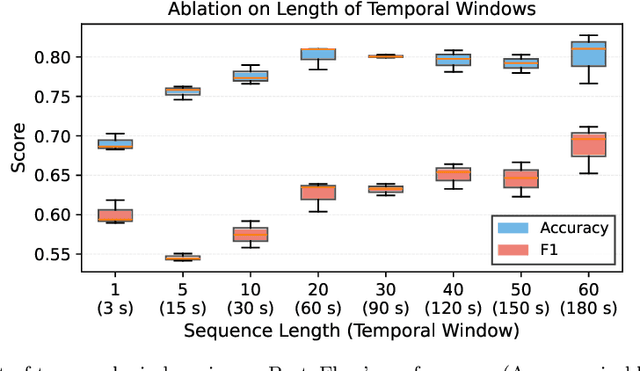
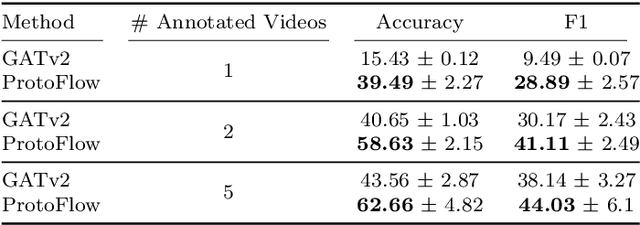
Abstract:Purpose: Detailed surgical recognition is critical for advancing AI-assisted surgery, yet progress is hampered by high annotation costs, data scarcity, and a lack of interpretable models. While scene graphs offer a structured abstraction of surgical events, their full potential remains untapped. In this work, we introduce ProtoFlow, a novel framework that learns dynamic scene graph prototypes to model complex surgical workflows in an interpretable and robust manner. Methods: ProtoFlow leverages a graph neural network (GNN) encoder-decoder architecture that combines self-supervised pretraining for rich representation learning with a prototype-based fine-tuning stage. This process discovers and refines core prototypes that encapsulate recurring, clinically meaningful patterns of surgical interaction, forming an explainable foundation for workflow analysis. Results: We evaluate our approach on the fine-grained CAT-SG dataset. ProtoFlow not only outperforms standard GNN baselines in overall accuracy but also demonstrates exceptional robustness in limited-data, few-shot scenarios, maintaining strong performance when trained on as few as one surgical video. Our qualitative analyses further show that the learned prototypes successfully identify distinct surgical sub-techniques and provide clear, interpretable insights into workflow deviations and rare complications. Conclusion: By uniting robust representation learning with inherent explainability, ProtoFlow represents a significant step toward developing more transparent, reliable, and data-efficient AI systems, accelerating their potential for clinical adoption in surgical training, real-time decision support, and workflow optimization.
UnReflectAnything: RGB-Only Highlight Removal by Rendering Synthetic Specular Supervision
Dec 11, 2025Abstract:Specular highlights distort appearance, obscure texture, and hinder geometric reasoning in both natural and surgical imagery. We present UnReflectAnything, an RGB-only framework that removes highlights from a single image by predicting a highlight map together with a reflection-free diffuse reconstruction. The model uses a frozen vision transformer encoder to extract multi-scale features, a lightweight head to localize specular regions, and a token-level inpainting module that restores corrupted feature patches before producing the final diffuse image. To overcome the lack of paired supervision, we introduce a Virtual Highlight Synthesis pipeline that renders physically plausible specularities using monocular geometry, Fresnel-aware shading, and randomized lighting which enables training on arbitrary RGB images with correct geometric structure. UnReflectAnything generalizes across natural and surgical domains where non-Lambertian surfaces and non-uniform lighting create severe highlights and it achieves competitive performance with state-of-the-art results on several benchmarks. Project Page: https://alberto-rota.github.io/UnReflectAnything/
LapFM: A Laparoscopic Segmentation Foundation Model via Hierarchical Concept Evolving Pre-training
Dec 09, 2025Abstract:Surgical segmentation is pivotal for scene understanding yet remains hindered by annotation scarcity and semantic inconsistency across diverse procedures. Existing approaches typically fine-tune natural foundation models (e.g., SAM) with limited supervision, functioning merely as domain adapters rather than surgical foundation models. Consequently, they struggle to generalize across the vast variability of surgical targets. To bridge this gap, we present LapFM, a foundation model designed to evolve robust segmentation capabilities from massive unlabeled surgical images. Distinct from medical foundation models relying on inefficient self-supervised proxy tasks, LapFM leverages a Hierarchical Concept Evolving Pre-training paradigm. First, we establish a Laparoscopic Concept Hierarchy (LCH) via a hierarchical mask decoder with parent-child query embeddings, unifying diverse entities (i.e., Anatomy, Tissue, and Instrument) into a scalable knowledge structure with cross-granularity semantic consistency. Second, we propose a Confidence-driven Evolving Labeling that iteratively generates and filters pseudo-labels based on hierarchical consistency, progressively incorporating reliable samples from unlabeled images into training. This process yields LapBench-114K, a large-scale benchmark comprising 114K image-mask pairs. Extensive experiments demonstrate that LapFM significantly outperforms state-of-the-art methods, establishing new standards for granularity-adaptive generalization in universal laparoscopic segmentation. The source code is available at https://github.com/xq141839/LapFM.
US-X Complete: A Multi-Modal Approach to Anatomical 3D Shape Recovery
Nov 19, 2025Abstract:Ultrasound offers a radiation-free, cost-effective solution for real-time visualization of spinal landmarks, paraspinal soft tissues and neurovascular structures, making it valuable for intraoperative guidance during spinal procedures. However, ultrasound suffers from inherent limitations in visualizing complete vertebral anatomy, in particular vertebral bodies, due to acoustic shadowing effects caused by bone. In this work, we present a novel multi-modal deep learning method for completing occluded anatomical structures in 3D ultrasound by leveraging complementary information from a single X-ray image. To enable training, we generate paired training data consisting of: (1) 2D lateral vertebral views that simulate X-ray scans, and (2) 3D partial vertebrae representations that mimic the limited visibility and occlusions encountered during ultrasound spine imaging. Our method integrates morphological information from both imaging modalities and demonstrates significant improvements in vertebral reconstruction (p < 0.001) compared to state of art in 3D ultrasound vertebral completion. We perform phantom studies as an initial step to future clinical translation, and achieve a more accurate, complete volumetric lumbar spine visualization overlayed on the ultrasound scan without the need for registration with preoperative modalities such as computed tomography. This demonstrates that integrating a single X-ray projection mitigates ultrasound's key limitation while preserving its strengths as the primary imaging modality. Code and data can be found at https://github.com/miruna20/US-X-Complete
From Linear Probing to Joint-Weighted Token Hierarchy: A Foundation Model Bridging Global and Cellular Representations in Biomarker Detection
Nov 07, 2025Abstract:AI-based biomarkers can infer molecular features directly from hematoxylin & eosin (H&E) slides, yet most pathology foundation models (PFMs) rely on global patch-level embeddings and overlook cell-level morphology. We present a PFM model, JWTH (Joint-Weighted Token Hierarchy), which integrates large-scale self-supervised pretraining with cell-centric post-tuning and attention pooling to fuse local and global tokens. Across four tasks involving four biomarkers and eight cohorts, JWTH achieves up to 8.3% higher balanced accuracy and 1.2% average improvement over prior PFMs, advancing interpretable and robust AI-based biomarker detection in digital pathology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge