Yuan Bi
Intelligent Virtual Sonographer (IVS): Enhancing Physician-Robot-Patient Communication
Jul 17, 2025Abstract:The advancement and maturity of large language models (LLMs) and robotics have unlocked vast potential for human-computer interaction, particularly in the field of robotic ultrasound. While existing research primarily focuses on either patient-robot or physician-robot interaction, the role of an intelligent virtual sonographer (IVS) bridging physician-robot-patient communication remains underexplored. This work introduces a conversational virtual agent in Extended Reality (XR) that facilitates real-time interaction between physicians, a robotic ultrasound system(RUS), and patients. The IVS agent communicates with physicians in a professional manner while offering empathetic explanations and reassurance to patients. Furthermore, it actively controls the RUS by executing physician commands and transparently relays these actions to the patient. By integrating LLM-powered dialogue with speech-to-text, text-to-speech, and robotic control, our system enhances the efficiency, clarity, and accessibility of robotic ultrasound acquisition. This work constitutes a first step toward understanding how IVS can bridge communication gaps in physician-robot-patient interaction, providing more control and therefore trust into physician-robot interaction while improving patient experience and acceptance of robotic ultrasound.
UltraAD: Fine-Grained Ultrasound Anomaly Classification via Few-Shot CLIP Adaptation
Jun 24, 2025Abstract:Precise anomaly detection in medical images is critical for clinical decision-making. While recent unsupervised or semi-supervised anomaly detection methods trained on large-scale normal data show promising results, they lack fine-grained differentiation, such as benign vs. malignant tumors. Additionally, ultrasound (US) imaging is highly sensitive to devices and acquisition parameter variations, creating significant domain gaps in the resulting US images. To address these challenges, we propose UltraAD, a vision-language model (VLM)-based approach that leverages few-shot US examples for generalized anomaly localization and fine-grained classification. To enhance localization performance, the image-level token of query visual prototypes is first fused with learnable text embeddings. This image-informed prompt feature is then further integrated with patch-level tokens, refining local representations for improved accuracy. For fine-grained classification, a memory bank is constructed from few-shot image samples and corresponding text descriptions that capture anatomical and abnormality-specific features. During training, the stored text embeddings remain frozen, while image features are adapted to better align with medical data. UltraAD has been extensively evaluated on three breast US datasets, outperforming state-of-the-art methods in both lesion localization and fine-grained medical classification. The code will be released upon acceptance.
Does Machine Unlearning Truly Remove Model Knowledge? A Framework for Auditing Unlearning in LLMs
May 29, 2025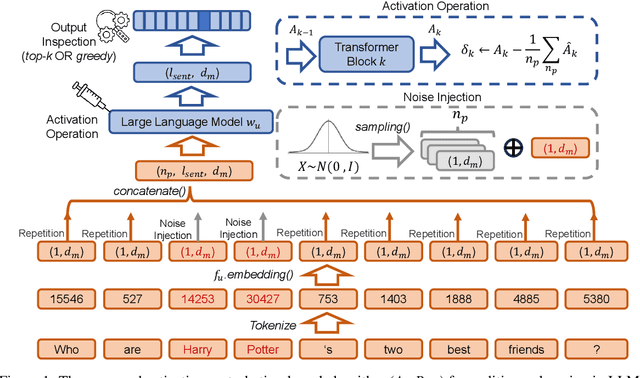
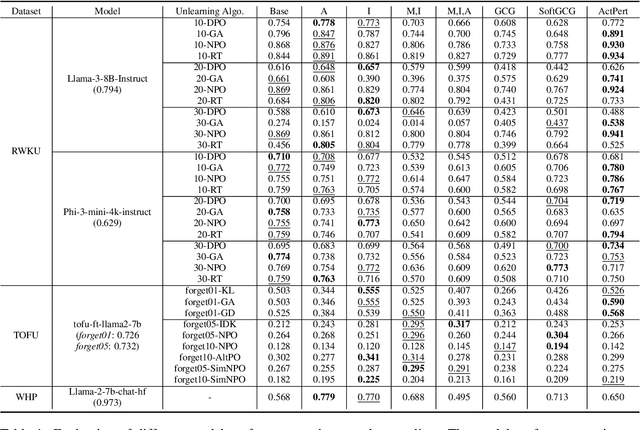
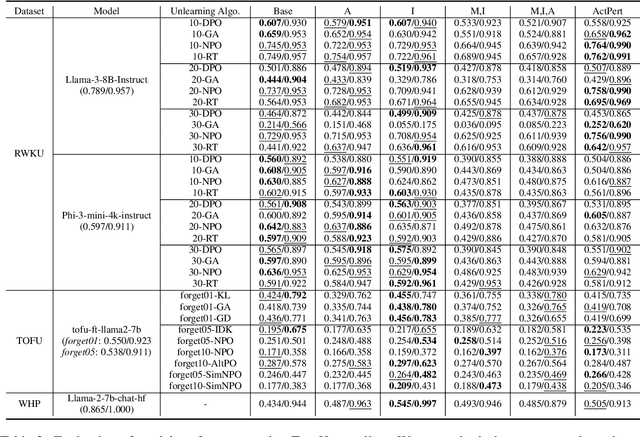
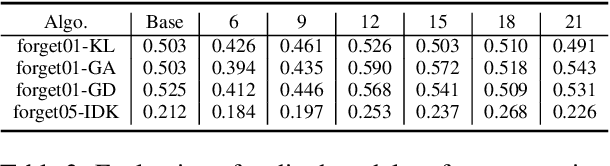
Abstract:In recent years, Large Language Models (LLMs) have achieved remarkable advancements, drawing significant attention from the research community. Their capabilities are largely attributed to large-scale architectures, which require extensive training on massive datasets. However, such datasets often contain sensitive or copyrighted content sourced from the public internet, raising concerns about data privacy and ownership. Regulatory frameworks, such as the General Data Protection Regulation (GDPR), grant individuals the right to request the removal of such sensitive information. This has motivated the development of machine unlearning algorithms that aim to remove specific knowledge from models without the need for costly retraining. Despite these advancements, evaluating the efficacy of unlearning algorithms remains a challenge due to the inherent complexity and generative nature of LLMs. In this work, we introduce a comprehensive auditing framework for unlearning evaluation, comprising three benchmark datasets, six unlearning algorithms, and five prompt-based auditing methods. By using various auditing algorithms, we evaluate the effectiveness and robustness of different unlearning strategies. To explore alternatives beyond prompt-based auditing, we propose a novel technique that leverages intermediate activation perturbations, addressing the limitations of auditing methods that rely solely on model inputs and outputs.
Robotic CBCT Meets Robotic Ultrasound
Feb 17, 2025Abstract:The multi-modality imaging system offers optimal fused images for safe and precise interventions in modern clinical practices, such as computed tomography - ultrasound (CT-US) guidance for needle insertion. However, the limited dexterity and mobility of current imaging devices hinder their integration into standardized workflows and the advancement toward fully autonomous intervention systems. In this paper, we present a novel clinical setup where robotic cone beam computed tomography (CBCT) and robotic US are pre-calibrated and dynamically co-registered, enabling new clinical applications. This setup allows registration-free rigid registration, facilitating multi-modal guided procedures in the absence of tissue deformation. First, a one-time pre-calibration is performed between the systems. To ensure a safe insertion path by highlighting critical vasculature on the 3D CBCT, SAM2 segments vessels from B-mode images, using the Doppler signal as an autonomously generated prompt. Based on the registration, the Doppler image or segmented vessel masks are then mapped onto the CBCT, creating an optimally fused image with comprehensive detail. To validate the system, we used a specially designed phantom, featuring lesions covered by ribs and multiple vessels with simulated moving flow. The mapping error between US and CBCT resulted in an average deviation of 1.72+-0.62 mm. A user study demonstrated the effectiveness of CBCT-US fusion for needle insertion guidance, showing significant improvements in time efficiency, accuracy, and success rate. Needle intervention performance improved by approximately 50% compared to the conventional US-guided workflow. We present the first robotic dual-modality imaging system designed to guide clinical applications. The results show significant performance improvements compared to traditional manual interventions.
Gaze-Guided Robotic Vascular Ultrasound Leveraging Human Intention Estimation
Feb 07, 2025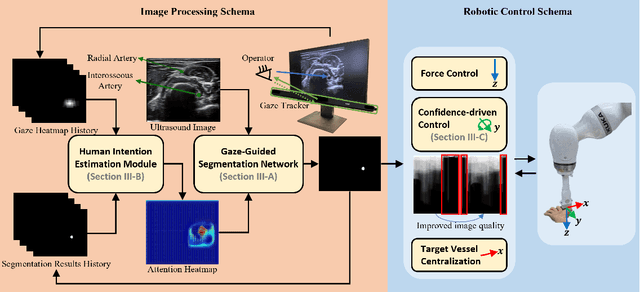
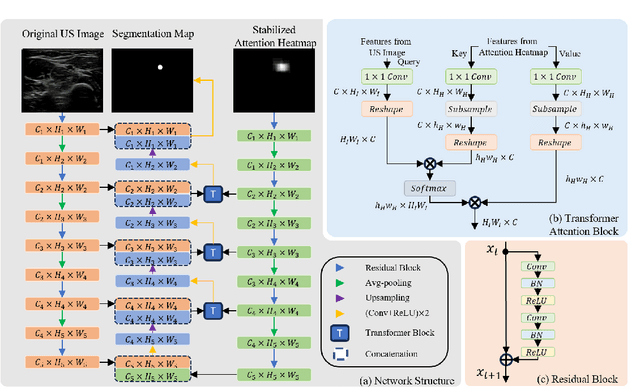
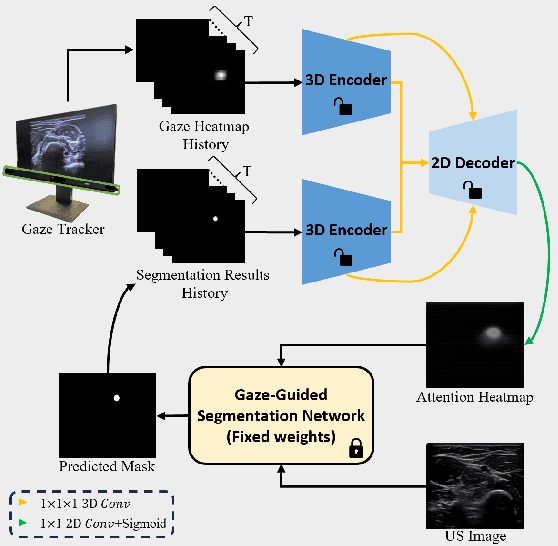
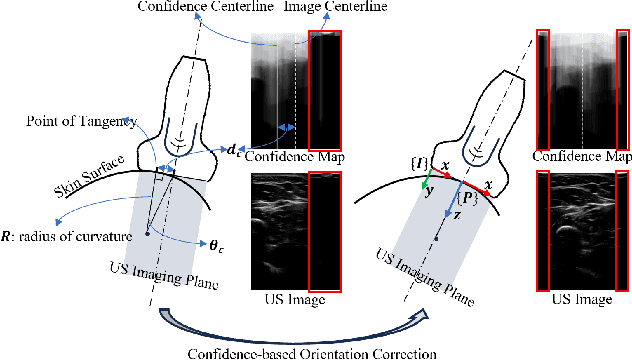
Abstract:Medical ultrasound has been widely used to examine vascular structure in modern clinical practice. However, traditional ultrasound examination often faces challenges related to inter- and intra-operator variation. The robotic ultrasound system (RUSS) appears as a potential solution for such challenges because of its superiority in stability and reproducibility. Given the complex anatomy of human vasculature, multiple vessels often appear in ultrasound images, or a single vessel bifurcates into branches, complicating the examination process. To tackle this challenge, this work presents a gaze-guided RUSS for vascular applications. A gaze tracker captures the eye movements of the operator. The extracted gaze signal guides the RUSS to follow the correct vessel when it bifurcates. Additionally, a gaze-guided segmentation network is proposed to enhance segmentation robustness by exploiting gaze information. However, gaze signals are often noisy, requiring interpretation to accurately discern the operator's true intentions. To this end, this study proposes a stabilization module to process raw gaze data. The inferred attention heatmap is utilized as a region proposal to aid segmentation and serve as a trigger signal when the operator needs to adjust the scanning target, such as when a bifurcation appears. To ensure appropriate contact between the probe and surface during scanning, an automatic ultrasound confidence-based orientation correction method is developed. In experiments, we demonstrated the efficiency of the proposed gaze-guided segmentation pipeline by comparing it with other methods. Besides, the performance of the proposed gaze-guided RUSS was also validated as a whole on a realistic arm phantom with an uneven surface.
Synomaly Noise and Multi-Stage Diffusion: A Novel Approach for Unsupervised Anomaly Detection in Ultrasound Imaging
Nov 06, 2024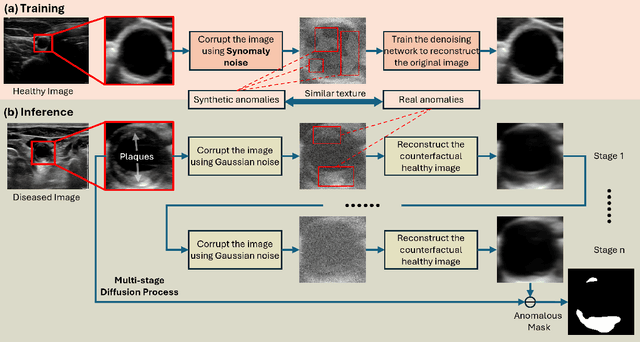
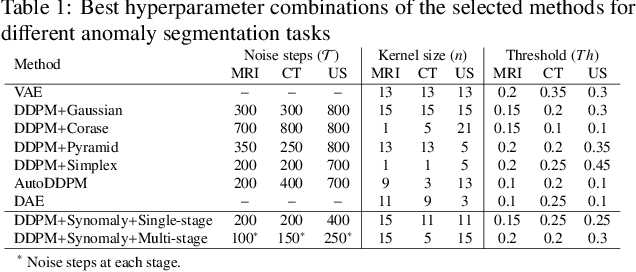
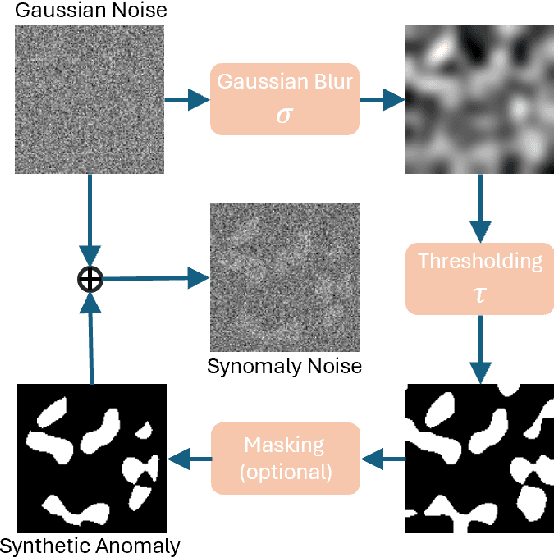
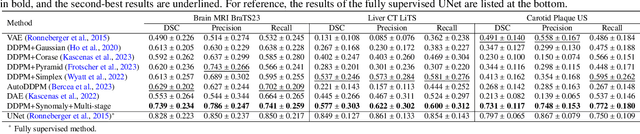
Abstract:Ultrasound (US) imaging is widely used in routine clinical practice due to its advantages of being radiation-free, cost-effective, and portable. However, the low reproducibility and quality of US images, combined with the scarcity of expert-level annotation, make the training of fully supervised segmentation models challenging. To address these issues, we propose a novel unsupervised anomaly detection framework based on a diffusion model that incorporates a synthetic anomaly (Synomaly) noise function and a multi-stage diffusion process. Synomaly noise introduces synthetic anomalies into healthy images during training, allowing the model to effectively learn anomaly removal. The multi-stage diffusion process is introduced to progressively denoise images, preserving fine details while improving the quality of anomaly-free reconstructions. The generated high-fidelity counterfactual healthy images can further enhance the interpretability of the segmentation models, as well as provide a reliable baseline for evaluating the extent of anomalies and supporting clinical decision-making. Notably, the unsupervised anomaly detection model is trained purely on healthy images, eliminating the need for anomalous training samples and pixel-level annotations. We validate the proposed approach on carotid US, brain MRI, and liver CT datasets. The experimental results demonstrate that the proposed framework outperforms existing state-of-the-art unsupervised anomaly detection methods, achieving performance comparable to fully supervised segmentation models in the US dataset. Additionally, ablation studies underline the importance of hyperparameter selection for Synomaly noise and the effectiveness of the multi-stage diffusion process in enhancing model performance.
Needle Segmentation Using GAN: Restoring Thin Instrument Visibility in Robotic Ultrasound
Jul 25, 2024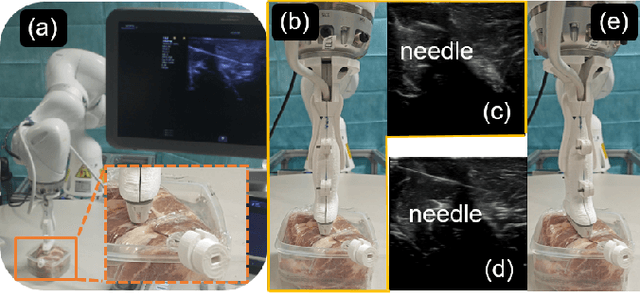
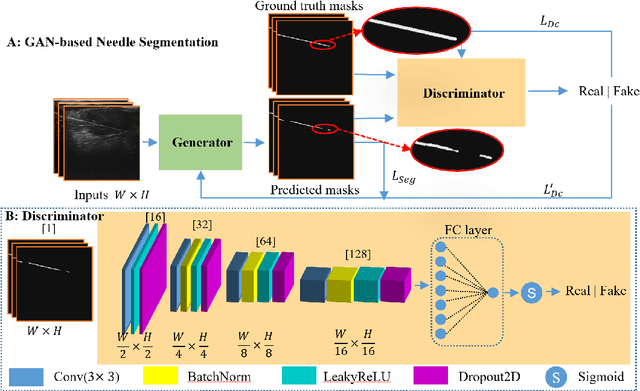
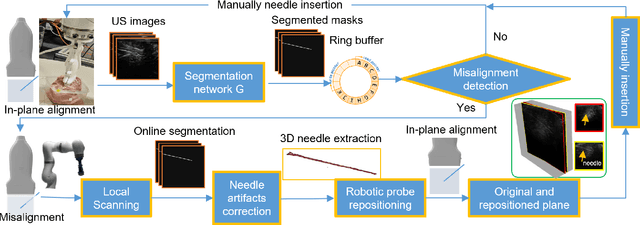
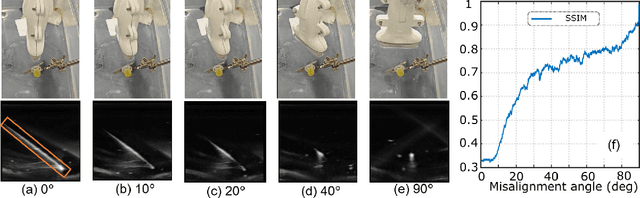
Abstract:Ultrasound-guided percutaneous needle insertion is a standard procedure employed in both biopsy and ablation in clinical practices. However, due to the complex interaction between tissue and instrument, the needle may deviate from the in-plane view, resulting in a lack of close monitoring of the percutaneous needle. To address this challenge, we introduce a robot-assisted ultrasound (US) imaging system designed to seamlessly monitor the insertion process and autonomously restore the visibility of the inserted instrument when misalignment happens. To this end, the adversarial structure is presented to encourage the generation of segmentation masks that align consistently with the ground truth in high-order space. This study also systematically investigates the effects on segmentation performance by exploring various training loss functions and their combinations. When misalignment between the probe and the percutaneous needle is detected, the robot is triggered to perform transverse searching to optimize the positional and rotational adjustment to restore needle visibility. The experimental results on ex-vivo porcine samples demonstrate that the proposed method can precisely segment the percutaneous needle (with a tip error of $0.37\pm0.29mm$ and an angle error of $1.19\pm 0.29^{\circ}$). Furthermore, the needle appearance can be successfully restored under the repositioned probe pose in all 45 trials, with repositioning errors of $1.51\pm0.95mm$ and $1.25\pm0.79^{\circ}$. from latex to text with math symbols
Class-Aware Cartilage Segmentation for Autonomous US-CT Registration in Robotic Intercostal Ultrasound Imaging
Jun 06, 2024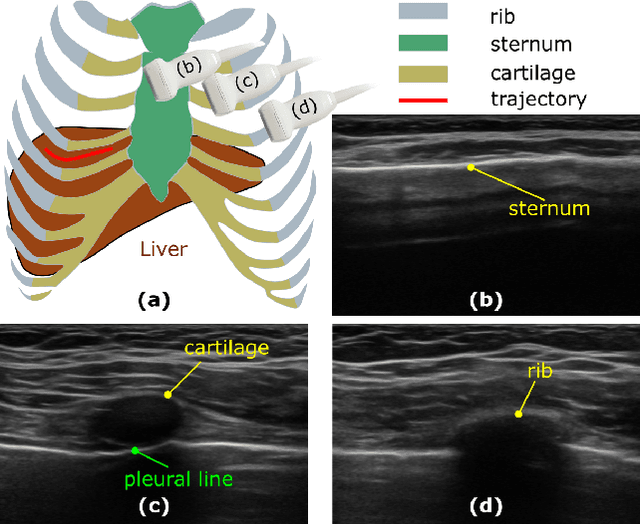
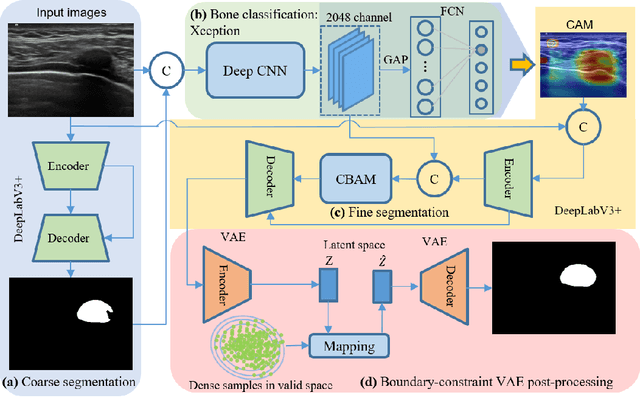
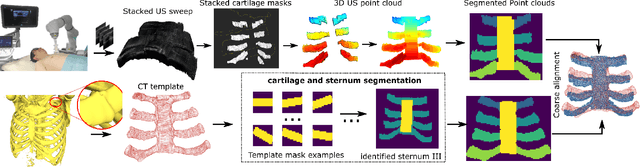
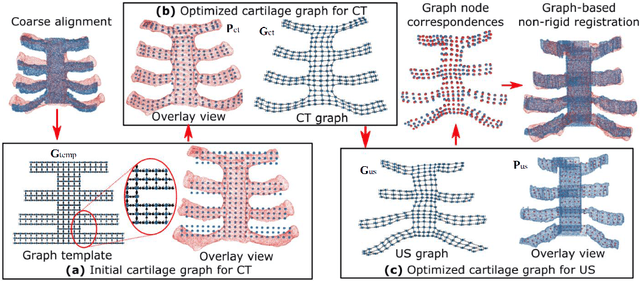
Abstract:Ultrasound imaging has been widely used in clinical examinations owing to the advantages of being portable, real-time, and radiation-free. Considering the potential of extensive deployment of autonomous examination systems in hospitals, robotic US imaging has attracted increased attention. However, due to the inter-patient variations, it is still challenging to have an optimal path for each patient, particularly for thoracic applications with limited acoustic windows, e.g., intercostal liver imaging. To address this problem, a class-aware cartilage bone segmentation network with geometry-constraint post-processing is presented to capture patient-specific rib skeletons. Then, a dense skeleton graph-based non-rigid registration is presented to map the intercostal scanning path from a generic template to individual patients. By explicitly considering the high-acoustic impedance bone structures, the transferred scanning path can be precisely located in the intercostal space, enhancing the visibility of internal organs by reducing the acoustic shadow. To evaluate the proposed approach, the final path mapping performance is validated on five distinct CTs and two volunteer US data, resulting in ten pairs of CT-US combinations. Results demonstrate that the proposed graph-based registration method can robustly and precisely map the path from CT template to individual patients (Euclidean error: $2.21\pm1.11~mm$).
Autonomous Path Planning for Intercostal Robotic Ultrasound Imaging Using Reinforcement Learning
Apr 15, 2024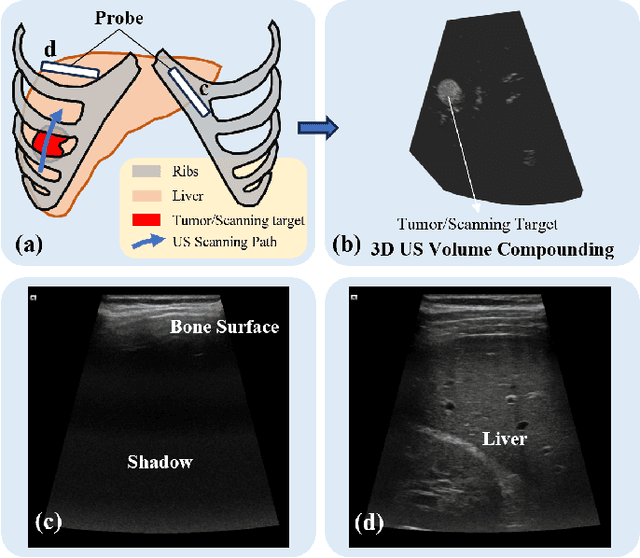
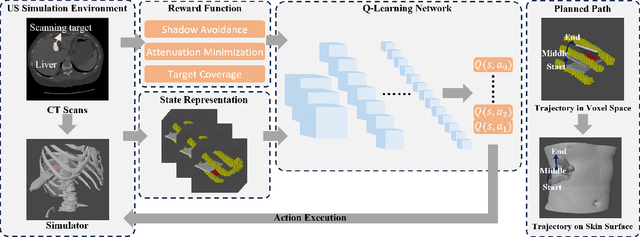
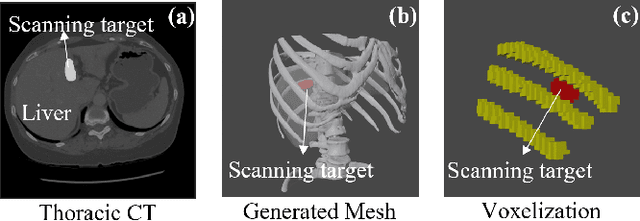
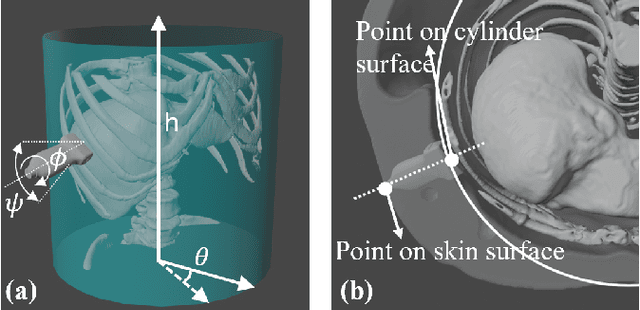
Abstract:Ultrasound (US) has been widely used in daily clinical practice for screening internal organs and guiding interventions. However, due to the acoustic shadow cast by the subcutaneous rib cage, the US examination for thoracic application is still challenging. To fully cover and reconstruct the region of interest in US for diagnosis, an intercostal scanning path is necessary. To tackle this challenge, we present a reinforcement learning (RL) approach for planning scanning paths between ribs to monitor changes in lesions on internal organs, such as the liver and heart, which are covered by rib cages. Structured anatomical information of the human skeleton is crucial for planning these intercostal paths. To obtain such anatomical insight, an RL agent is trained in a virtual environment constructed using computational tomography (CT) templates with randomly initialized tumors of various shapes and locations. In addition, task-specific state representation and reward functions are introduced to ensure the convergence of the training process while minimizing the effects of acoustic attenuation and shadows during scanning. To validate the effectiveness of the proposed approach, experiments have been carried out on unseen CTs with randomly defined single or multiple scanning targets. The results demonstrate the efficiency of the proposed RL framework in planning non-shadowed US scanning trajectories in areas with limited acoustic access.
Machine Learning in Robotic Ultrasound Imaging: Challenges and Perspectives
Jan 04, 2024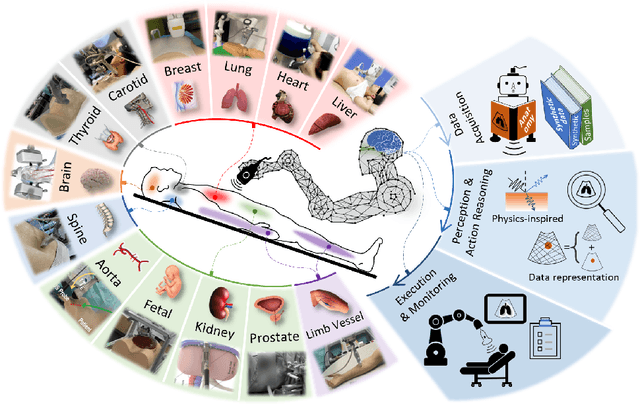
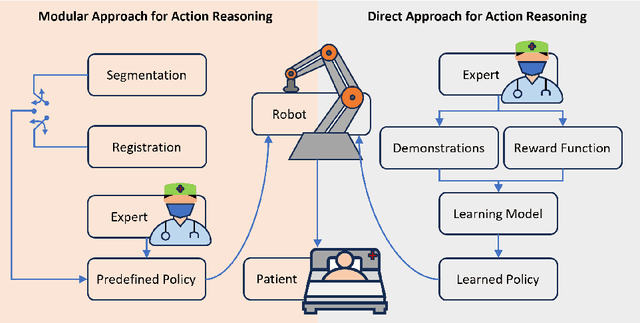
Abstract:This article reviews the recent advances in intelligent robotic ultrasound (US) imaging systems. We commence by presenting the commonly employed robotic mechanisms and control techniques in robotic US imaging, along with their clinical applications. Subsequently, we focus on the deployment of machine learning techniques in the development of robotic sonographers, emphasizing crucial developments aimed at enhancing the intelligence of these systems. The methods for achieving autonomous action reasoning are categorized into two sets of approaches: those relying on implicit environmental data interpretation and those using explicit interpretation. Throughout this exploration, we also discuss practical challenges, including those related to the scarcity of medical data, the need for a deeper understanding of the physical aspects involved, and effective data representation approaches. Moreover, we conclude by highlighting the open problems in the field and analyzing different possible perspectives on how the community could move forward in this research area.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge