Lucie Huang
Synomaly Noise and Multi-Stage Diffusion: A Novel Approach for Unsupervised Anomaly Detection in Ultrasound Imaging
Nov 06, 2024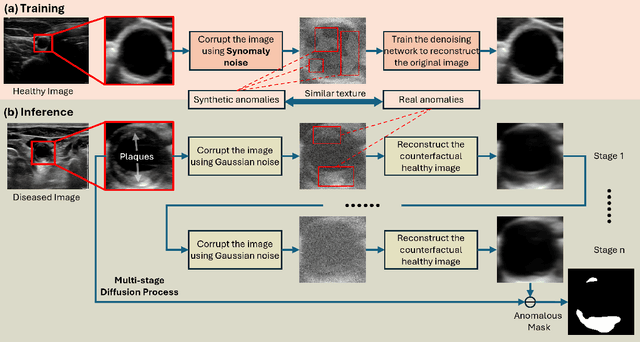
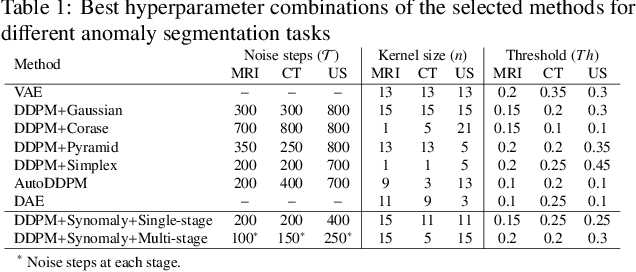
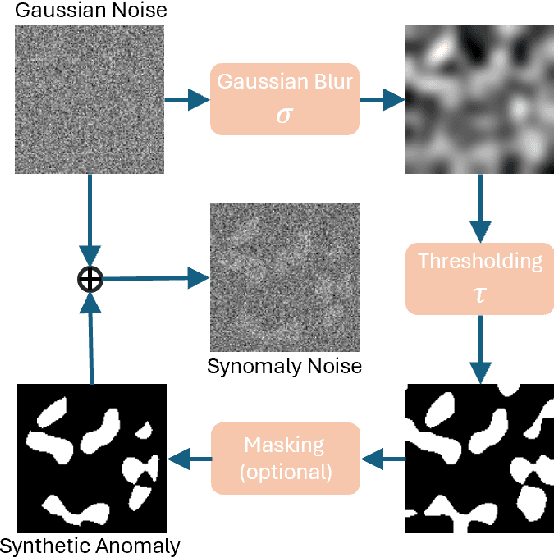
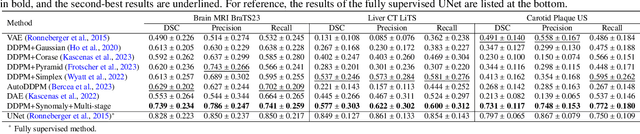
Abstract:Ultrasound (US) imaging is widely used in routine clinical practice due to its advantages of being radiation-free, cost-effective, and portable. However, the low reproducibility and quality of US images, combined with the scarcity of expert-level annotation, make the training of fully supervised segmentation models challenging. To address these issues, we propose a novel unsupervised anomaly detection framework based on a diffusion model that incorporates a synthetic anomaly (Synomaly) noise function and a multi-stage diffusion process. Synomaly noise introduces synthetic anomalies into healthy images during training, allowing the model to effectively learn anomaly removal. The multi-stage diffusion process is introduced to progressively denoise images, preserving fine details while improving the quality of anomaly-free reconstructions. The generated high-fidelity counterfactual healthy images can further enhance the interpretability of the segmentation models, as well as provide a reliable baseline for evaluating the extent of anomalies and supporting clinical decision-making. Notably, the unsupervised anomaly detection model is trained purely on healthy images, eliminating the need for anomalous training samples and pixel-level annotations. We validate the proposed approach on carotid US, brain MRI, and liver CT datasets. The experimental results demonstrate that the proposed framework outperforms existing state-of-the-art unsupervised anomaly detection methods, achieving performance comparable to fully supervised segmentation models in the US dataset. Additionally, ablation studies underline the importance of hyperparameter selection for Synomaly noise and the effectiveness of the multi-stage diffusion process in enhancing model performance.
Outlier Detection using Self-Organizing Maps for Automated Blood Cell Analysis
Aug 18, 2022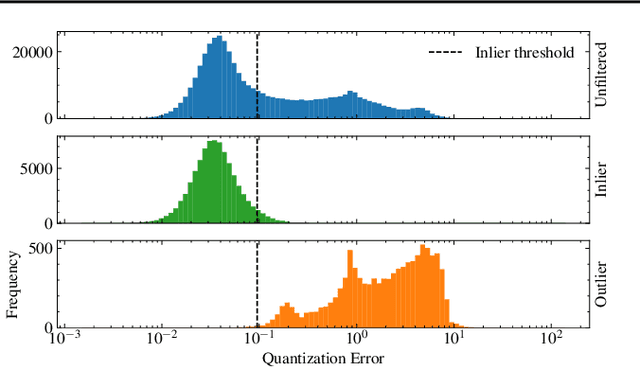
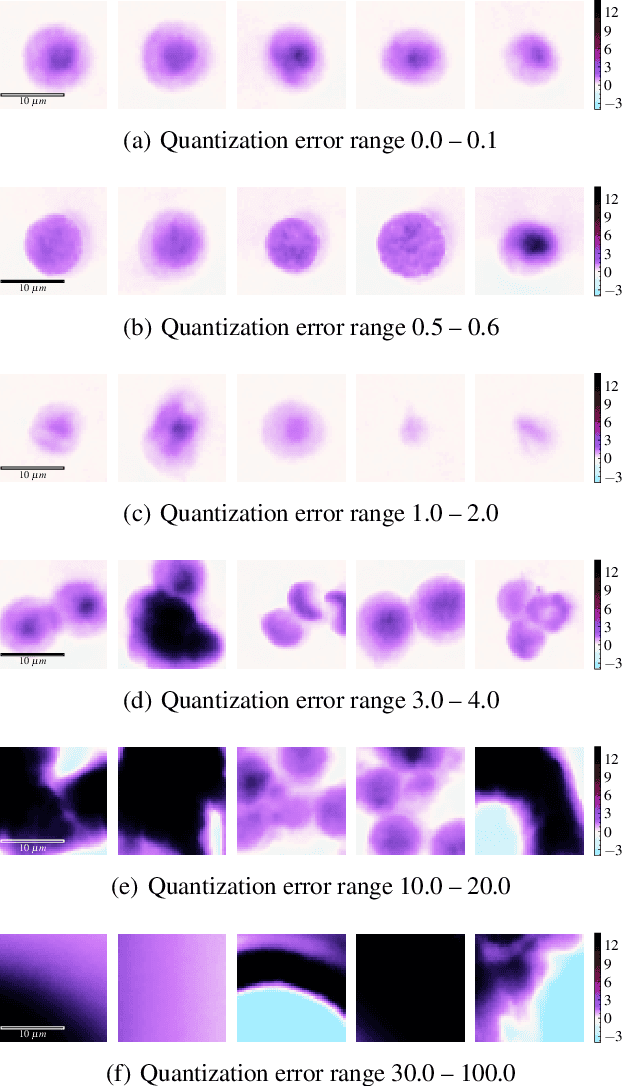
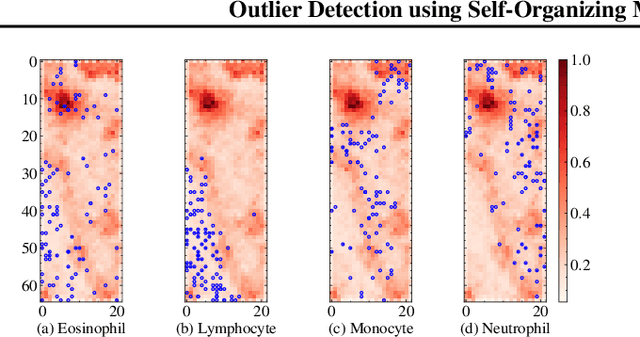
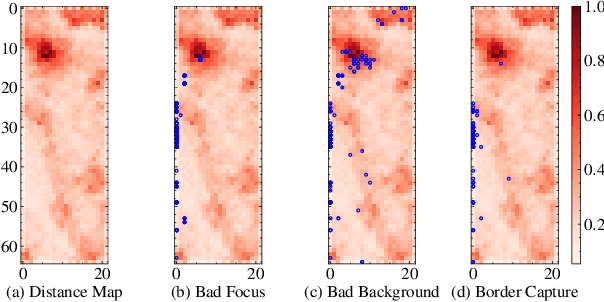
Abstract:The quality of datasets plays a crucial role in the successful training and deployment of deep learning models. Especially in the medical field, where system performance may impact the health of patients, clean datasets are a safety requirement for reliable predictions. Therefore, outlier detection is an essential process when building autonomous clinical decision systems. In this work, we assess the suitability of Self-Organizing Maps for outlier detection specifically on a medical dataset containing quantitative phase images of white blood cells. We detect and evaluate outliers based on quantization errors and distance maps. Our findings confirm the suitability of Self-Organizing Maps for unsupervised Out-Of-Distribution detection on the dataset at hand. Self-Organizing Maps perform on par with a manually specified filter based on expert domain knowledge. Additionally, they show promise as a tool in the exploration and cleaning of medical datasets. As a direction for future research, we suggest a combination of Self-Organizing Maps and feature extraction based on deep learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge