Mohammad Farid Azampour
US-X Complete: A Multi-Modal Approach to Anatomical 3D Shape Recovery
Nov 19, 2025Abstract:Ultrasound offers a radiation-free, cost-effective solution for real-time visualization of spinal landmarks, paraspinal soft tissues and neurovascular structures, making it valuable for intraoperative guidance during spinal procedures. However, ultrasound suffers from inherent limitations in visualizing complete vertebral anatomy, in particular vertebral bodies, due to acoustic shadowing effects caused by bone. In this work, we present a novel multi-modal deep learning method for completing occluded anatomical structures in 3D ultrasound by leveraging complementary information from a single X-ray image. To enable training, we generate paired training data consisting of: (1) 2D lateral vertebral views that simulate X-ray scans, and (2) 3D partial vertebrae representations that mimic the limited visibility and occlusions encountered during ultrasound spine imaging. Our method integrates morphological information from both imaging modalities and demonstrates significant improvements in vertebral reconstruction (p < 0.001) compared to state of art in 3D ultrasound vertebral completion. We perform phantom studies as an initial step to future clinical translation, and achieve a more accurate, complete volumetric lumbar spine visualization overlayed on the ultrasound scan without the need for registration with preoperative modalities such as computed tomography. This demonstrates that integrating a single X-ray projection mitigates ultrasound's key limitation while preserving its strengths as the primary imaging modality. Code and data can be found at https://github.com/miruna20/US-X-Complete
UltrON: Ultrasound Occupancy Networks
Sep 10, 2025Abstract:In free-hand ultrasound imaging, sonographers rely on expertise to mentally integrate partial 2D views into 3D anatomical shapes. Shape reconstruction can assist clinicians in this process. Central to this task is the choice of shape representation, as it determines how accurately and efficiently the structure can be visualized, analyzed, and interpreted. Implicit representations, such as SDF and occupancy function, offer a powerful alternative to traditional voxel- or mesh-based methods by modeling continuous, smooth surfaces with compact storage, avoiding explicit discretization. Recent studies demonstrate that SDF can be effectively optimized using annotations derived from segmented B-mode ultrasound images. Yet, these approaches hinge on precise annotations, overlooking the rich acoustic information embedded in B-mode intensity. Moreover, implicit representation approaches struggle with the ultrasound's view-dependent nature and acoustic shadowing artifacts, which impair reconstruction. To address the problems resulting from occlusions and annotation dependency, we propose an occupancy-based representation and introduce \gls{UltrON} that leverages acoustic features to improve geometric consistency in weakly-supervised optimization regime. We show that these features can be obtained from B-mode images without additional annotation cost. Moreover, we propose a novel loss function that compensates for view-dependency in the B-mode images and facilitates occupancy optimization from multiview ultrasound. By incorporating acoustic properties, \gls{UltrON} generalizes to shapes of the same anatomy. We show that \gls{UltrON} mitigates the limitations of occlusions and sparse labeling and paves the way for more accurate 3D reconstruction. Code and dataset will be available at https://github.com/magdalena-wysocki/ultron.
Shape Completion and Real-Time Visualization in Robotic Ultrasound Spine Acquisitions
Aug 12, 2025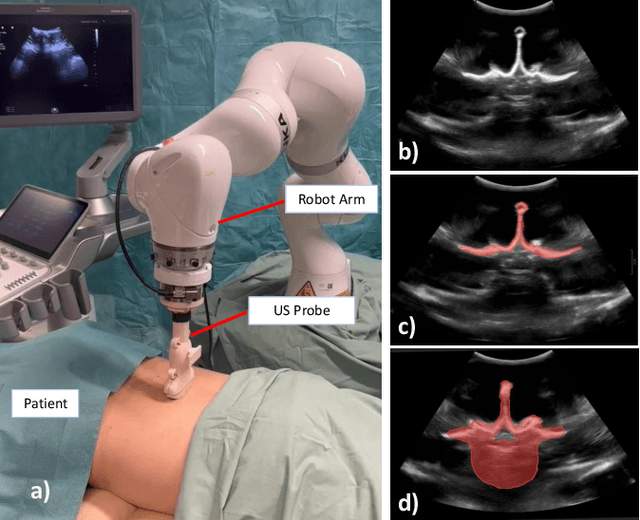
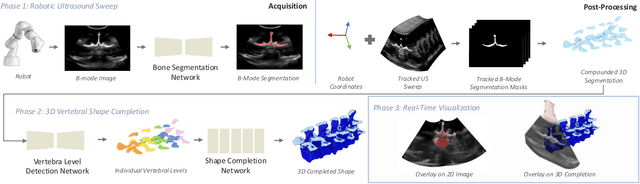
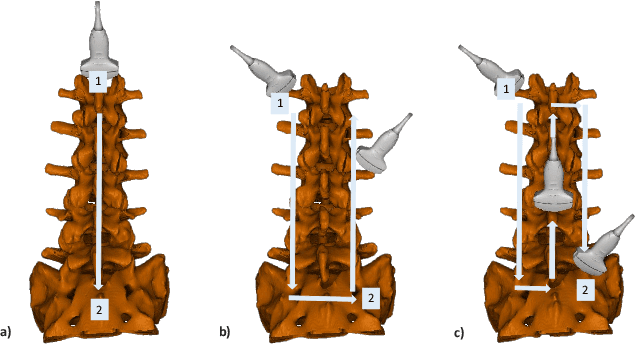
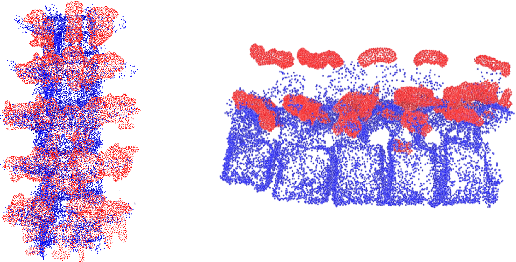
Abstract:Ultrasound (US) imaging is increasingly used in spinal procedures due to its real-time, radiation-free capabilities; however, its effectiveness is hindered by shadowing artifacts that obscure deeper tissue structures. Traditional approaches, such as CT-to-US registration, incorporate anatomical information from preoperative CT scans to guide interventions, but they are limited by complex registration requirements, differences in spine curvature, and the need for recent CT imaging. Recent shape completion methods can offer an alternative by reconstructing spinal structures in US data, while being pretrained on large set of publicly available CT scans. However, these approaches are typically offline and have limited reproducibility. In this work, we introduce a novel integrated system that combines robotic ultrasound with real-time shape completion to enhance spinal visualization. Our robotic platform autonomously acquires US sweeps of the lumbar spine, extracts vertebral surfaces from ultrasound, and reconstructs the complete anatomy using a deep learning-based shape completion network. This framework provides interactive, real-time visualization with the capability to autonomously repeat scans and can enable navigation to target locations. This can contribute to better consistency, reproducibility, and understanding of the underlying anatomy. We validate our approach through quantitative experiments assessing shape completion accuracy and evaluations of multiple spine acquisition protocols on a phantom setup. Additionally, we present qualitative results of the visualization on a volunteer scan.
HyperSORT: Self-Organising Robust Training with hyper-networks
Jun 26, 2025Abstract:Medical imaging datasets often contain heterogeneous biases ranging from erroneous labels to inconsistent labeling styles. Such biases can negatively impact deep segmentation networks performance. Yet, the identification and characterization of such biases is a particularly tedious and challenging task. In this paper, we introduce HyperSORT, a framework using a hyper-network predicting UNets' parameters from latent vectors representing both the image and annotation variability. The hyper-network parameters and the latent vector collection corresponding to each data sample from the training set are jointly learned. Hence, instead of optimizing a single neural network to fit a dataset, HyperSORT learns a complex distribution of UNet parameters where low density areas can capture noise-specific patterns while larger modes robustly segment organs in differentiated but meaningful manners. We validate our method on two 3D abdominal CT public datasets: first a synthetically perturbed version of the AMOS dataset, and TotalSegmentator, a large scale dataset containing real unknown biases and errors. Our experiments show that HyperSORT creates a structured mapping of the dataset allowing the identification of relevant systematic biases and erroneous samples. Latent space clusters yield UNet parameters performing the segmentation task in accordance with the underlying learned systematic bias. The code and our analysis of the TotalSegmentator dataset are made available: https://github.com/ImFusionGmbH/HyperSORT
Skelite: Compact Neural Networks for Efficient Iterative Skeletonization
Mar 10, 2025



Abstract:Skeletonization extracts thin representations from images that compactly encode their geometry and topology. These representations have become an important topological prior for preserving connectivity in curvilinear structures, aiding medical tasks like vessel segmentation. Existing compatible skeletonization algorithms face significant trade-offs: morphology-based approaches are computationally efficient but prone to frequent breakages, while topology-preserving methods require substantial computational resources. We propose a novel framework for training iterative skeletonization algorithms with a learnable component. The framework leverages synthetic data, task-specific augmentation, and a model distillation strategy to learn compact neural networks that produce thin, connected skeletons with a fully differentiable iterative algorithm. Our method demonstrates a 100 times speedup over topology-constrained algorithms while maintaining high accuracy and generalizing effectively to new domains without fine-tuning. Benchmarking and downstream validation in 2D and 3D tasks demonstrate its computational efficiency and real-world applicability
UltraRay: Full-Path Ray Tracing for Enhancing Realism in Ultrasound Simulation
Jan 10, 2025



Abstract:Traditional ultrasound simulators solve the wave equation to model pressure distribution fields, achieving high accuracy but requiring significant computational time and resources. To address this, ray tracing approaches have been introduced, modeling wave propagation as rays interacting with boundaries and scatterers. However, existing models simplify ray propagation, generating echoes at interaction points without considering return paths to the sensor. This can result in unrealistic artifacts and necessitates careful scene tuning for plausible results. We propose a novel ultrasound simulation pipeline that utilizes a ray tracing algorithm to generate echo data, tracing each ray from the transducer through the scene and back to the sensor. To replicate advanced ultrasound imaging, we introduce a ray emission scheme optimized for plane wave imaging, incorporating delay and steering capabilities. Furthermore, we integrate a standard signal processing pipeline to simulate end-to-end ultrasound image formation. We showcase the efficacy of the proposed pipeline by modeling synthetic scenes featuring highly reflective objects, such as bones. In doing so, our proposed approach, UltraRay, not only enhances the overall visual quality but also improves the realism of the simulated images by accurately capturing secondary reflections and reducing unnatural artifacts. By building on top of a differentiable framework, the proposed pipeline lays the groundwork for a fast and differentiable ultrasound simulation tool necessary for gradient-based optimization, enabling advanced ultrasound beamforming strategies, neural network integration, and accurate inverse scene reconstruction.
Intraoperative Registration by Cross-Modal Inverse Neural Rendering
Sep 18, 2024Abstract:We present in this paper a novel approach for 3D/2D intraoperative registration during neurosurgery via cross-modal inverse neural rendering. Our approach separates implicit neural representation into two components, handling anatomical structure preoperatively and appearance intraoperatively. This disentanglement is achieved by controlling a Neural Radiance Field's appearance with a multi-style hypernetwork. Once trained, the implicit neural representation serves as a differentiable rendering engine, which can be used to estimate the surgical camera pose by minimizing the dissimilarity between its rendered images and the target intraoperative image. We tested our method on retrospective patients' data from clinical cases, showing that our method outperforms state-of-the-art while meeting current clinical standards for registration. Code and additional resources can be found at https://maxfehrentz.github.io/style-ngp/.
PHOCUS: Physics-Based Deconvolution for Ultrasound Resolution Enhancement
Aug 07, 2024Abstract:Ultrasound is widely used in medical diagnostics allowing for accessible and powerful imaging but suffers from resolution limitations due to diffraction and the finite aperture of the imaging system, which restricts diagnostic use. The impulse function of an ultrasound imaging system is called the point spread function (PSF), which is convolved with the spatial distribution of reflectors in the image formation process. Recovering high-resolution reflector distributions by removing image distortions induced by the convolution process improves image clarity and detail. Conventionally, deconvolution techniques attempt to rectify the imaging system's dependent PSF, working directly on the radio-frequency (RF) data. However, RF data is often not readily accessible. Therefore, we introduce a physics-based deconvolution process using a modeled PSF, working directly on the more commonly available B-mode images. By leveraging Implicit Neural Representations (INRs), we learn a continuous mapping from spatial locations to their respective echogenicity values, effectively compensating for the discretized image space. Our contribution consists of a novel methodology for retrieving a continuous echogenicity map directly from a B-mode image through a differentiable physics-based rendering pipeline for ultrasound resolution enhancement. We qualitatively and quantitatively evaluate our approach on synthetic data, demonstrating improvements over traditional methods in metrics such as PSNR and SSIM. Furthermore, we show qualitative enhancements on an ultrasound phantom and an in-vivo acquisition of a carotid artery.
Diffusion as Sound Propagation: Physics-inspired Model for Ultrasound Image Generation
Jul 07, 2024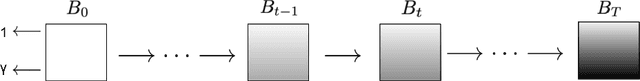
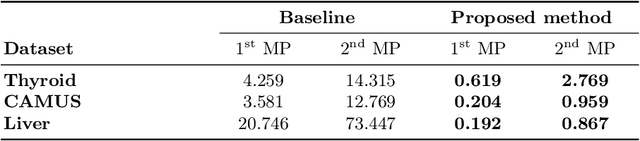
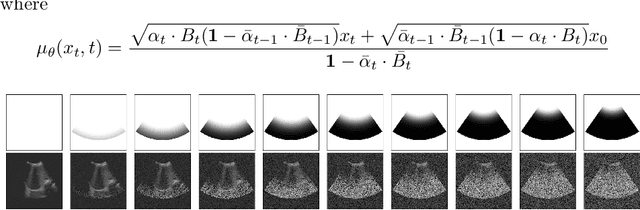
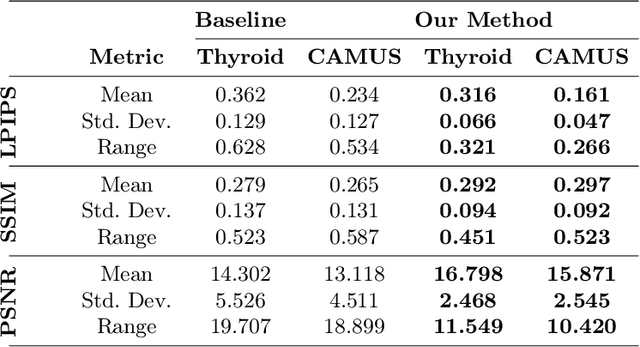
Abstract:Deep learning (DL) methods typically require large datasets to effectively learn data distributions. However, in the medical field, data is often limited in quantity, and acquiring labeled data can be costly. To mitigate this data scarcity, data augmentation techniques are commonly employed. Among these techniques, generative models play a pivotal role in expanding datasets. However, when it comes to ultrasound (US) imaging, the authenticity of generated data often diminishes due to the oversight of ultrasound physics. We propose a novel approach to improve the quality of generated US images by introducing a physics-based diffusion model that is specifically designed for this image modality. The proposed model incorporates an US-specific scheduler scheme that mimics the natural behavior of sound wave propagation in ultrasound imaging. Our analysis demonstrates how the proposed method aids in modeling the attenuation dynamics in US imaging. We present both qualitative and quantitative results based on standard generative model metrics, showing that our proposed method results in overall more plausible images. Our code is available at https://github.com/marinadominguez/diffusion-for-us-images
Shape Completion in the Dark: Completing Vertebrae Morphology from 3D Ultrasound
Apr 11, 2024Abstract:Purpose: Ultrasound (US) imaging, while advantageous for its radiation-free nature, is challenging to interpret due to only partially visible organs and a lack of complete 3D information. While performing US-based diagnosis or investigation, medical professionals therefore create a mental map of the 3D anatomy. In this work, we aim to replicate this process and enhance the visual representation of anatomical structures. Methods: We introduce a point-cloud-based probabilistic DL method to complete occluded anatomical structures through 3D shape completion and choose US-based spine examinations as our application. To enable training, we generate synthetic 3D representations of partially occluded spinal views by mimicking US physics and accounting for inherent artifacts. Results: The proposed model performs consistently on synthetic and patient data, with mean and median differences of 2.02 and 0.03 in CD, respectively. Our ablation study demonstrates the importance of US physics-based data generation, reflected in the large mean and median difference of 11.8 CD and 9.55 CD, respectively. Additionally, we demonstrate that anatomic landmarks, such as the spinous process (with reconstruction CD of 4.73) and the facet joints (mean distance to GT of 4.96mm) are preserved in the 3D completion. Conclusion: Our work establishes the feasibility of 3D shape completion for lumbar vertebrae, ensuring the preservation of level-wise characteristics and successful generalization from synthetic to real data. The incorporation of US physics contributes to more accurate patient data completions. Notably, our method preserves essential anatomic landmarks and reconstructs crucial injections sites at their correct locations. The generated data and source code will be made publicly available (https://github.com/miruna20/Shape-Completion-in-the-Dark).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge