Mahdi Saleh
ConceptPose: Training-Free Zero-Shot Object Pose Estimation using Concept Vectors
Dec 19, 2025Abstract:Object pose estimation is a fundamental task in computer vision and robotics, yet most methods require extensive, dataset-specific training. Concurrently, large-scale vision language models show remarkable zero-shot capabilities. In this work, we bridge these two worlds by introducing ConceptPose, a framework for object pose estimation that is both training-free and model-free. ConceptPose leverages a vision-language-model (VLM) to create open-vocabulary 3D concept maps, where each point is tagged with a concept vector derived from saliency maps. By establishing robust 3D-3D correspondences across concept maps, our approach allows precise estimation of 6DoF relative pose. Without any object or dataset-specific training, our approach achieves state-of-the-art results on common zero shot relative pose estimation benchmarks, significantly outperforming existing methods by over 62% in ADD(-S) score, including those that utilize extensive dataset-specific training.
PRISM: Probabilistic Representation for Integrated Shape Modeling and Generation
Apr 06, 2025Abstract:Despite the advancements in 3D full-shape generation, accurately modeling complex geometries and semantics of shape parts remains a significant challenge, particularly for shapes with varying numbers of parts. Current methods struggle to effectively integrate the contextual and structural information of 3D shapes into their generative processes. We address these limitations with PRISM, a novel compositional approach for 3D shape generation that integrates categorical diffusion models with Statistical Shape Models (SSM) and Gaussian Mixture Models (GMM). Our method employs compositional SSMs to capture part-level geometric variations and uses GMM to represent part semantics in a continuous space. This integration enables both high fidelity and diversity in generated shapes while preserving structural coherence. Through extensive experiments on shape generation and manipulation tasks, we demonstrate that our approach significantly outperforms previous methods in both quality and controllability of part-level operations. Our code will be made publicly available.
ESCAPE: Equivariant Shape Completion via Anchor Point Encoding
Dec 01, 2024Abstract:Shape completion, a crucial task in 3D computer vision, involves predicting and filling the missing regions of scanned or partially observed objects. Current methods expect known pose or canonical coordinates and do not perform well under varying rotations, limiting their real-world applicability. We introduce ESCAPE (Equivariant Shape Completion via Anchor Point Encoding), a novel framework designed to achieve rotation-equivariant shape completion. Our approach employs a distinctive encoding strategy by selecting anchor points from a shape and representing all points as a distance to all anchor points. This enables the model to capture a consistent, rotation-equivariant understanding of the object's geometry. ESCAPE leverages a transformer architecture to encode and decode the distance transformations, ensuring that generated shape completions remain accurate and equivariant under rotational transformations. Subsequently, we perform optimization to calculate the predicted shapes from the encodings. Experimental evaluations demonstrate that ESCAPE achieves robust, high-quality reconstructions across arbitrary rotations and translations, showcasing its effectiveness in real-world applications without additional pose estimation modules.
Deep Spectral Methods for Unsupervised Ultrasound Image Interpretation
Aug 04, 2024Abstract:Ultrasound imaging is challenging to interpret due to non-uniform intensities, low contrast, and inherent artifacts, necessitating extensive training for non-specialists. Advanced representation with clear tissue structure separation could greatly assist clinicians in mapping underlying anatomy and distinguishing between tissue layers. Decomposing an image into semantically meaningful segments is mainly achieved using supervised segmentation algorithms. Unsupervised methods are beneficial, as acquiring large labeled datasets is difficult and costly, but despite their advantages, they still need to be explored in ultrasound. This paper proposes a novel unsupervised deep learning strategy tailored to ultrasound to obtain easily interpretable tissue separations. We integrate key concepts from unsupervised deep spectral methods, which combine spectral graph theory with deep learning methods. We utilize self-supervised transformer features for spectral clustering to generate meaningful segments based on ultrasound-specific metrics and shape and positional priors, ensuring semantic consistency across the dataset. We evaluate our unsupervised deep learning strategy on three ultrasound datasets, showcasing qualitative results across anatomical contexts without label requirements. We also conduct a comparative analysis against other clustering algorithms to demonstrate superior segmentation performance, boundary preservation, and label consistency.
Shape Completion in the Dark: Completing Vertebrae Morphology from 3D Ultrasound
Apr 11, 2024Abstract:Purpose: Ultrasound (US) imaging, while advantageous for its radiation-free nature, is challenging to interpret due to only partially visible organs and a lack of complete 3D information. While performing US-based diagnosis or investigation, medical professionals therefore create a mental map of the 3D anatomy. In this work, we aim to replicate this process and enhance the visual representation of anatomical structures. Methods: We introduce a point-cloud-based probabilistic DL method to complete occluded anatomical structures through 3D shape completion and choose US-based spine examinations as our application. To enable training, we generate synthetic 3D representations of partially occluded spinal views by mimicking US physics and accounting for inherent artifacts. Results: The proposed model performs consistently on synthetic and patient data, with mean and median differences of 2.02 and 0.03 in CD, respectively. Our ablation study demonstrates the importance of US physics-based data generation, reflected in the large mean and median difference of 11.8 CD and 9.55 CD, respectively. Additionally, we demonstrate that anatomic landmarks, such as the spinous process (with reconstruction CD of 4.73) and the facet joints (mean distance to GT of 4.96mm) are preserved in the 3D completion. Conclusion: Our work establishes the feasibility of 3D shape completion for lumbar vertebrae, ensuring the preservation of level-wise characteristics and successful generalization from synthetic to real data. The incorporation of US physics contributes to more accurate patient data completions. Notably, our method preserves essential anatomic landmarks and reconstructs crucial injections sites at their correct locations. The generated data and source code will be made publicly available (https://github.com/miruna20/Shape-Completion-in-the-Dark).
Physics-Encoded Graph Neural Networks for Deformation Prediction under Contact
Feb 05, 2024



Abstract:In robotics, it's crucial to understand object deformation during tactile interactions. A precise understanding of deformation can elevate robotic simulations and have broad implications across different industries. We introduce a method using Physics-Encoded Graph Neural Networks (GNNs) for such predictions. Similar to robotic grasping and manipulation scenarios, we focus on modeling the dynamics between a rigid mesh contacting a deformable mesh under external forces. Our approach represents both the soft body and the rigid body within graph structures, where nodes hold the physical states of the meshes. We also incorporate cross-attention mechanisms to capture the interplay between the objects. By jointly learning geometry and physics, our model reconstructs consistent and detailed deformations. We've made our code and dataset public to advance research in robotic simulation and grasping.
Dynamic Hyperbolic Attention Network for Fine Hand-object Reconstruction
Sep 06, 2023Abstract:Reconstructing both objects and hands in 3D from a single RGB image is complex. Existing methods rely on manually defined hand-object constraints in Euclidean space, leading to suboptimal feature learning. Compared with Euclidean space, hyperbolic space better preserves the geometric properties of meshes thanks to its exponentially-growing space distance, which amplifies the differences between the features based on similarity. In this work, we propose the first precise hand-object reconstruction method in hyperbolic space, namely Dynamic Hyperbolic Attention Network (DHANet), which leverages intrinsic properties of hyperbolic space to learn representative features. Our method that projects mesh and image features into a unified hyperbolic space includes two modules, ie. dynamic hyperbolic graph convolution and image-attention hyperbolic graph convolution. With these two modules, our method learns mesh features with rich geometry-image multi-modal information and models better hand-object interaction. Our method provides a promising alternative for fine hand-object reconstruction in hyperbolic space. Extensive experiments on three public datasets demonstrate that our method outperforms most state-of-the-art methods.
On the Localization of Ultrasound Image Slices within Point Distribution Models
Sep 01, 2023Abstract:Thyroid disorders are most commonly diagnosed using high-resolution Ultrasound (US). Longitudinal nodule tracking is a pivotal diagnostic protocol for monitoring changes in pathological thyroid morphology. This task, however, imposes a substantial cognitive load on clinicians due to the inherent challenge of maintaining a mental 3D reconstruction of the organ. We thus present a framework for automated US image slice localization within a 3D shape representation to ease how such sonographic diagnoses are carried out. Our proposed method learns a common latent embedding space between US image patches and the 3D surface of an individual's thyroid shape, or a statistical aggregation in the form of a statistical shape model (SSM), via contrastive metric learning. Using cross-modality registration and Procrustes analysis, we leverage features from our model to register US slices to a 3D mesh representation of the thyroid shape. We demonstrate that our multi-modal registration framework can localize images on the 3D surface topology of a patient-specific organ and the mean shape of an SSM. Experimental results indicate slice positions can be predicted within an average of 1.2 mm of the ground-truth slice location on the patient-specific 3D anatomy and 4.6 mm on the SSM, exemplifying its usefulness for slice localization during sonographic acquisitions. Code is publically available: \href{https://github.com/vuenc/slice-to-shape}{https://github.com/vuenc/slice-to-shape}
S3M: Scalable Statistical Shape Modeling through Unsupervised Correspondences
Apr 15, 2023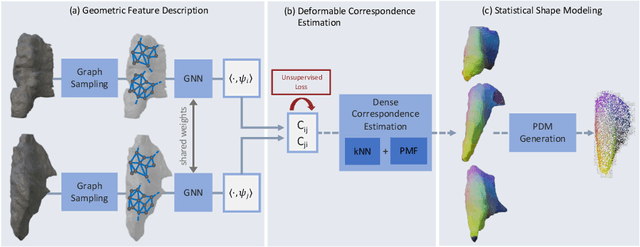
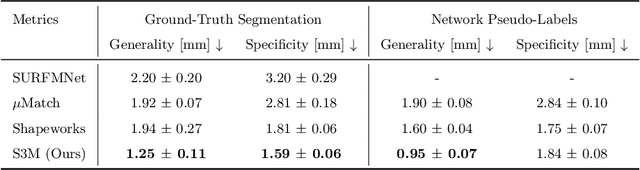
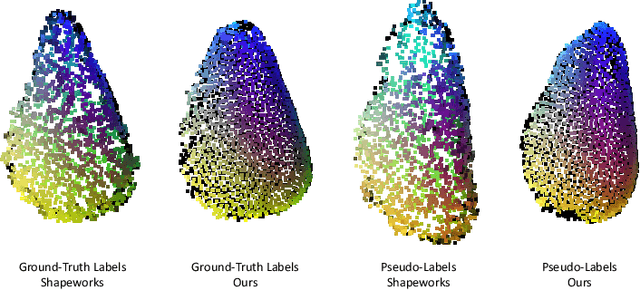
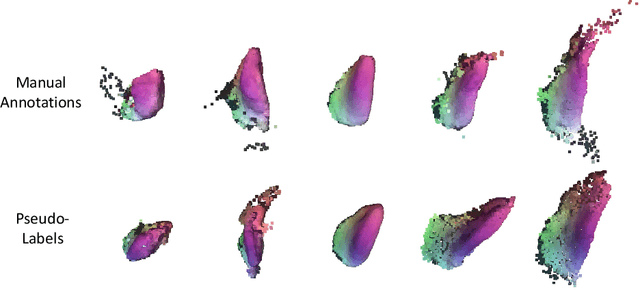
Abstract:Statistical shape models (SSMs) are an established way to geometrically represent the anatomy of a population with various clinically relevant applications. However, they typically require domain expertise and labor-intensive manual segmentations or landmark annotations to generate. Methods to estimate correspondences for SSMs typically learn with such labels as supervision signals. We address these shortcomings by proposing an unsupervised method that leverages deep geometric features and functional correspondences to learn local and global shape structures across complex anatomies simultaneously. Our pipeline significantly improves unsupervised correspondence estimation for SSMs compared to baseline methods, even on highly irregular surface topologies. We demonstrate this for two different anatomical structures: the thyroid and a multi-chamber heart dataset. Furthermore, our method is robust enough to learn from noisy neural network predictions, enabling scaling SSMs to larger patient populations without manual annotation.
Rotation-Invariant Transformer for Point Cloud Matching
Mar 25, 2023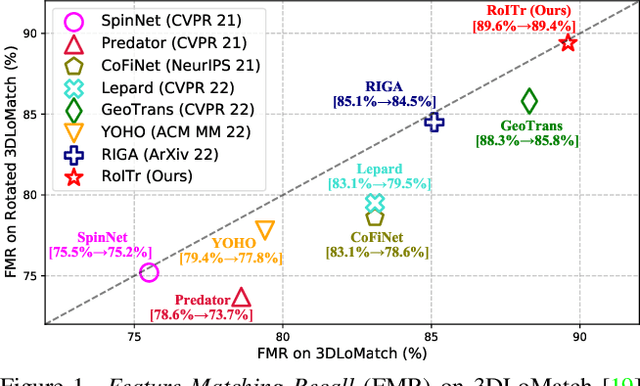
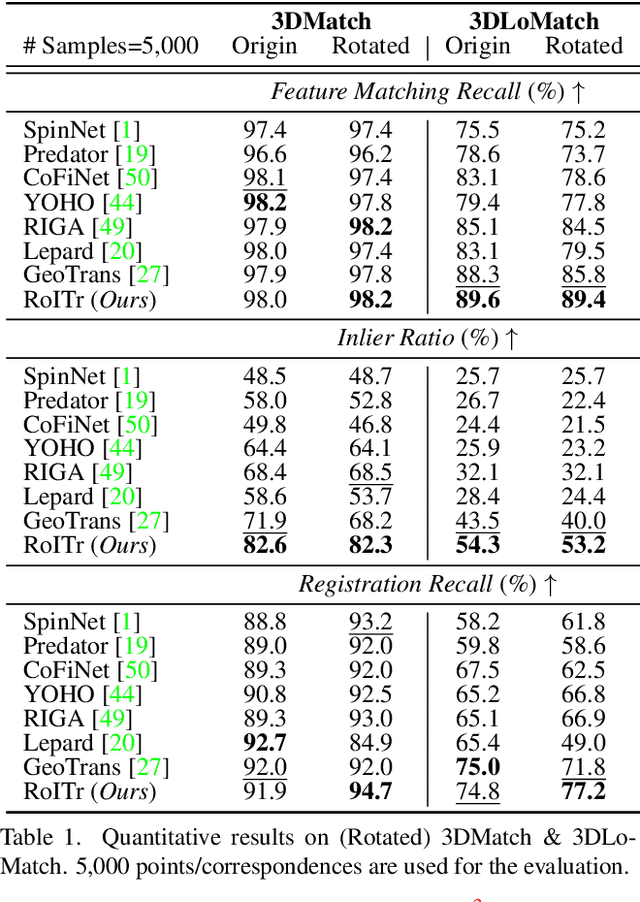
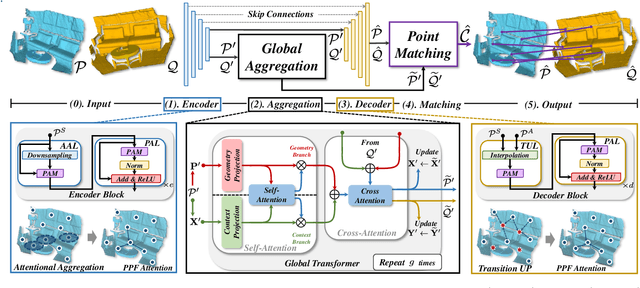
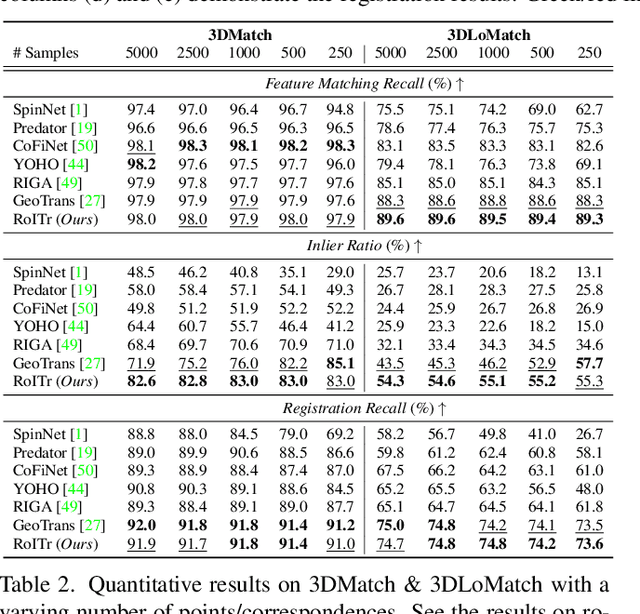
Abstract:The intrinsic rotation invariance lies at the core of matching point clouds with handcrafted descriptors. However, it is widely despised by recent deep matchers that obtain the rotation invariance extrinsically via data augmentation. As the finite number of augmented rotations can never span the continuous SO(3) space, these methods usually show instability when facing rotations that are rarely seen. To this end, we introduce RoITr, a Rotation-Invariant Transformer to cope with the pose variations in the point cloud matching task. We contribute both on the local and global levels. Starting from the local level, we introduce an attention mechanism embedded with Point Pair Feature (PPF)-based coordinates to describe the pose-invariant geometry, upon which a novel attention-based encoder-decoder architecture is constructed. We further propose a global transformer with rotation-invariant cross-frame spatial awareness learned by the self-attention mechanism, which significantly improves the feature distinctiveness and makes the model robust with respect to the low overlap. Experiments are conducted on both the rigid and non-rigid public benchmarks, where RoITr outperforms all the state-of-the-art models by a considerable margin in the low-overlapping scenarios. Especially when the rotations are enlarged on the challenging 3DLoMatch benchmark, RoITr surpasses the existing methods by at least 13 and 5 percentage points in terms of Inlier Ratio and Registration Recall, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge