Alexander Bauman
S3M: Scalable Statistical Shape Modeling through Unsupervised Correspondences
Apr 15, 2023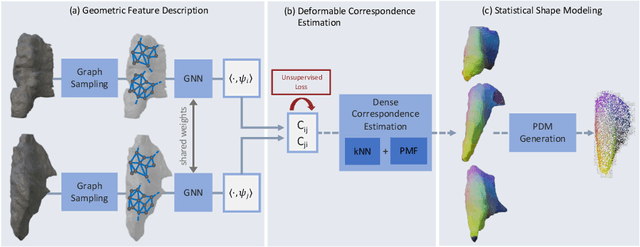
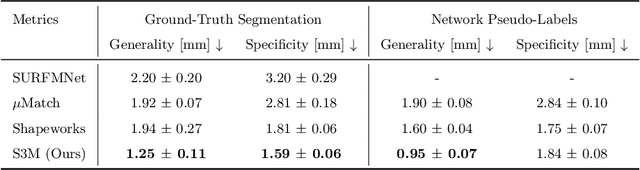
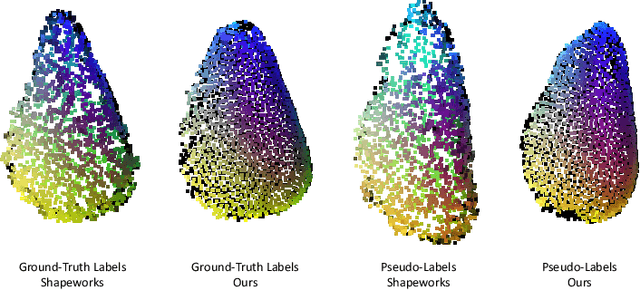
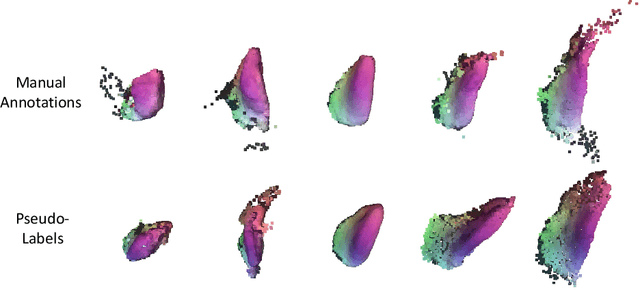
Abstract:Statistical shape models (SSMs) are an established way to geometrically represent the anatomy of a population with various clinically relevant applications. However, they typically require domain expertise and labor-intensive manual segmentations or landmark annotations to generate. Methods to estimate correspondences for SSMs typically learn with such labels as supervision signals. We address these shortcomings by proposing an unsupervised method that leverages deep geometric features and functional correspondences to learn local and global shape structures across complex anatomies simultaneously. Our pipeline significantly improves unsupervised correspondence estimation for SSMs compared to baseline methods, even on highly irregular surface topologies. We demonstrate this for two different anatomical structures: the thyroid and a multi-chamber heart dataset. Furthermore, our method is robust enough to learn from noisy neural network predictions, enabling scaling SSMs to larger patient populations without manual annotation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge