Vincent Bürgin
Efficient Betti Matching Enables Topology-Aware 3D Segmentation via Persistent Homology
Jul 05, 2024Abstract:In this work, we propose an efficient algorithm for the calculation of the Betti matching, which can be used as a loss function to train topology aware segmentation networks. Betti matching loss builds on techniques from topological data analysis, specifically persistent homology. A major challenge is the computational cost of computing persistence barcodes. In response to this challenge, we propose a new, highly optimized implementation of Betti matching, implemented in C++ together with a python interface, which achieves significant speedups compared to the state-of-the-art implementation Cubical Ripser. We use Betti matching 3D to train segmentation networks with the Betti matching loss and demonstrate improved topological correctness of predicted segmentations across several datasets. The source code is available at https://github.com/nstucki/Betti-Matching-3D.
On the Localization of Ultrasound Image Slices within Point Distribution Models
Sep 01, 2023Abstract:Thyroid disorders are most commonly diagnosed using high-resolution Ultrasound (US). Longitudinal nodule tracking is a pivotal diagnostic protocol for monitoring changes in pathological thyroid morphology. This task, however, imposes a substantial cognitive load on clinicians due to the inherent challenge of maintaining a mental 3D reconstruction of the organ. We thus present a framework for automated US image slice localization within a 3D shape representation to ease how such sonographic diagnoses are carried out. Our proposed method learns a common latent embedding space between US image patches and the 3D surface of an individual's thyroid shape, or a statistical aggregation in the form of a statistical shape model (SSM), via contrastive metric learning. Using cross-modality registration and Procrustes analysis, we leverage features from our model to register US slices to a 3D mesh representation of the thyroid shape. We demonstrate that our multi-modal registration framework can localize images on the 3D surface topology of a patient-specific organ and the mean shape of an SSM. Experimental results indicate slice positions can be predicted within an average of 1.2 mm of the ground-truth slice location on the patient-specific 3D anatomy and 4.6 mm on the SSM, exemplifying its usefulness for slice localization during sonographic acquisitions. Code is publically available: \href{https://github.com/vuenc/slice-to-shape}{https://github.com/vuenc/slice-to-shape}
Robust vertebra identification using simultaneous node and edge predicting Graph Neural Networks
Jul 27, 2023Abstract:Automatic vertebra localization and identification in CT scans is important for numerous clinical applications. Much progress has been made on this topic, but it mostly targets positional localization of vertebrae, ignoring their orientation. Additionally, most methods employ heuristics in their pipeline that can be sensitive in real clinical images which tend to contain abnormalities. We introduce a simple pipeline that employs a standard prediction with a U-Net, followed by a single graph neural network to associate and classify vertebrae with full orientation. To test our method, we introduce a new vertebra dataset that also contains pedicle detections that are associated with vertebra bodies, creating a more challenging landmark prediction, association and classification task. Our method is able to accurately associate the correct body and pedicle landmarks, ignore false positives and classify vertebrae in a simple, fully trainable pipeline avoiding application-specific heuristics. We show our method outperforms traditional approaches such as Hungarian Matching and Hidden Markov Models. We also show competitive performance on the standard VerSe challenge body identification task.
S3M: Scalable Statistical Shape Modeling through Unsupervised Correspondences
Apr 15, 2023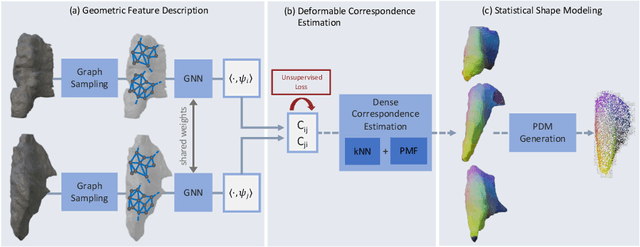
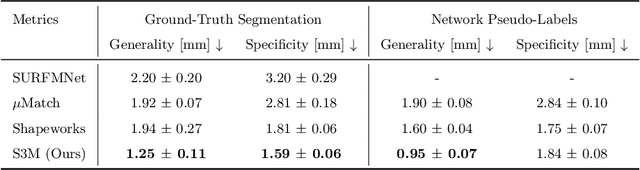
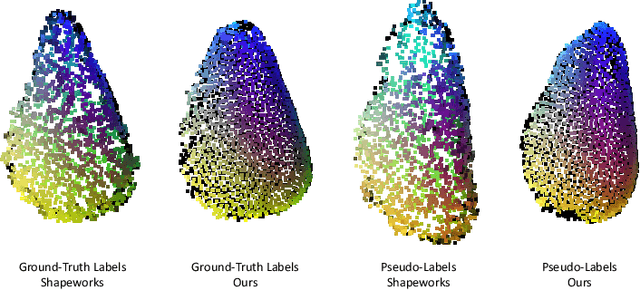
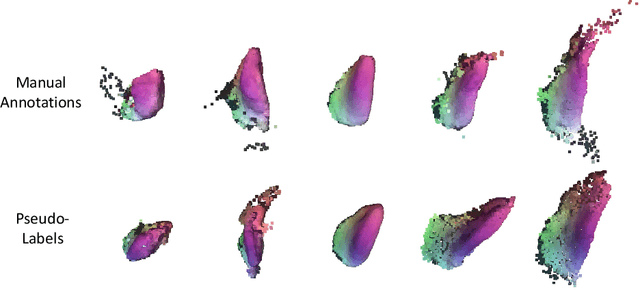
Abstract:Statistical shape models (SSMs) are an established way to geometrically represent the anatomy of a population with various clinically relevant applications. However, they typically require domain expertise and labor-intensive manual segmentations or landmark annotations to generate. Methods to estimate correspondences for SSMs typically learn with such labels as supervision signals. We address these shortcomings by proposing an unsupervised method that leverages deep geometric features and functional correspondences to learn local and global shape structures across complex anatomies simultaneously. Our pipeline significantly improves unsupervised correspondence estimation for SSMs compared to baseline methods, even on highly irregular surface topologies. We demonstrate this for two different anatomical structures: the thyroid and a multi-chamber heart dataset. Furthermore, our method is robust enough to learn from noisy neural network predictions, enabling scaling SSMs to larger patient populations without manual annotation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge