Thomas Wendler
Beyond the LUMIR challenge: The pathway to foundational registration models
May 30, 2025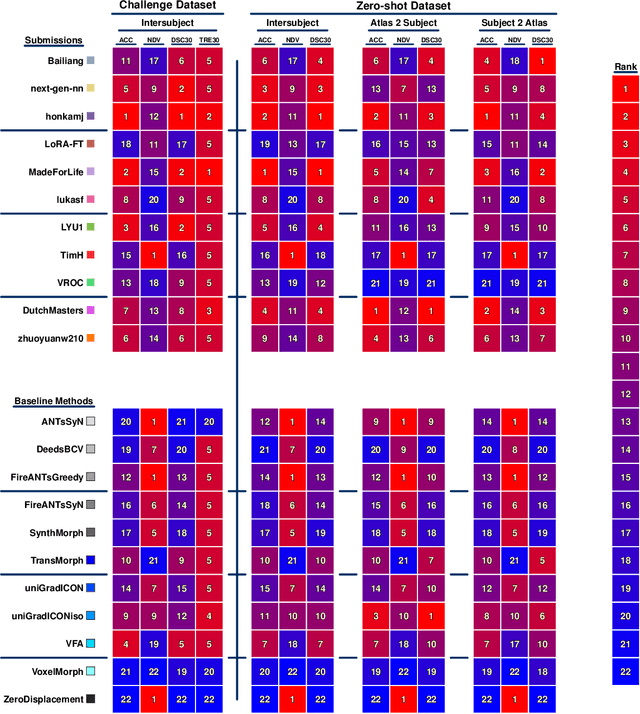
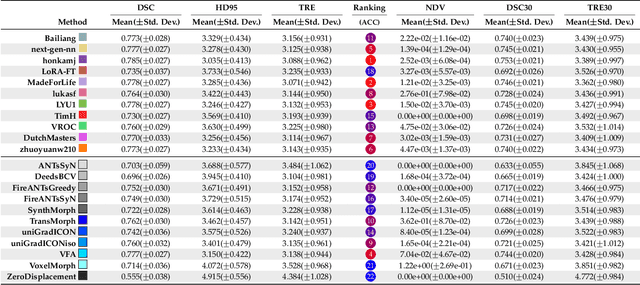
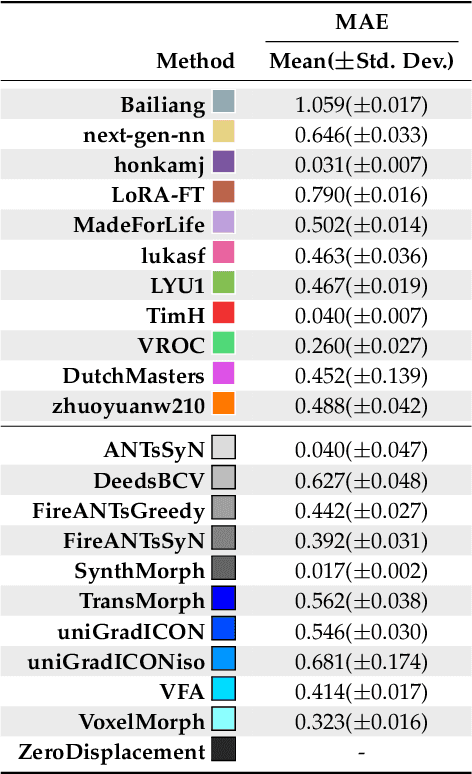
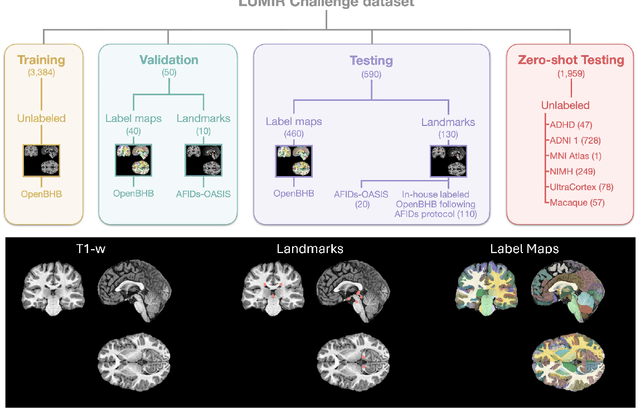
Abstract:Medical image challenges have played a transformative role in advancing the field, catalyzing algorithmic innovation and establishing new performance standards across diverse clinical applications. Image registration, a foundational task in neuroimaging pipelines, has similarly benefited from the Learn2Reg initiative. Building on this foundation, we introduce the Large-scale Unsupervised Brain MRI Image Registration (LUMIR) challenge, a next-generation benchmark designed to assess and advance unsupervised brain MRI registration. Distinct from prior challenges that leveraged anatomical label maps for supervision, LUMIR removes this dependency by providing over 4,000 preprocessed T1-weighted brain MRIs for training without any label maps, encouraging biologically plausible deformation modeling through self-supervision. In addition to evaluating performance on 590 held-out test subjects, LUMIR introduces a rigorous suite of zero-shot generalization tasks, spanning out-of-domain imaging modalities (e.g., FLAIR, T2-weighted, T2*-weighted), disease populations (e.g., Alzheimer's disease), acquisition protocols (e.g., 9.4T MRI), and species (e.g., macaque brains). A total of 1,158 subjects and over 4,000 image pairs were included for evaluation. Performance was assessed using both segmentation-based metrics (Dice coefficient, 95th percentile Hausdorff distance) and landmark-based registration accuracy (target registration error). Across both in-domain and zero-shot tasks, deep learning-based methods consistently achieved state-of-the-art accuracy while producing anatomically plausible deformation fields. The top-performing deep learning-based models demonstrated diffeomorphic properties and inverse consistency, outperforming several leading optimization-based methods, and showing strong robustness to most domain shifts, the exception being a drop in performance on out-of-domain contrasts.
Physiological neural representation for personalised tracer kinetic parameter estimation from dynamic PET
Apr 23, 2025Abstract:Dynamic positron emission tomography (PET) with [$^{18}$F]FDG enables non-invasive quantification of glucose metabolism through kinetic analysis, often modelled by the two-tissue compartment model (TCKM). However, voxel-wise kinetic parameter estimation using conventional methods is computationally intensive and limited by spatial resolution. Deep neural networks (DNNs) offer an alternative but require large training datasets and significant computational resources. To address these limitations, we propose a physiological neural representation based on implicit neural representations (INRs) for personalized kinetic parameter estimation. INRs, which learn continuous functions, allow for efficient, high-resolution parametric imaging with reduced data requirements. Our method also integrates anatomical priors from a 3D CT foundation model to enhance robustness and precision in kinetic modelling. We evaluate our approach on an [$^{18}$F]FDG dynamic PET/CT dataset and compare it to state-of-the-art DNNs. Results demonstrate superior spatial resolution, lower mean-squared error, and improved anatomical consistency, particularly in tumour and highly vascularized regions. Our findings highlight the potential of INRs for personalized, data-efficient tracer kinetic modelling, enabling applications in tumour characterization, segmentation, and prognostic assessment.
Anatomy-constrained modelling of image-derived input functions in dynamic PET using multi-organ segmentation
Apr 23, 2025Abstract:Accurate kinetic analysis of [$^{18}$F]FDG distribution in dynamic positron emission tomography (PET) requires anatomically constrained modelling of image-derived input functions (IDIFs). Traditionally, IDIFs are obtained from the aorta, neglecting anatomical variations and complex vascular contributions. This study proposes a multi-organ segmentation-based approach that integrates IDIFs from the aorta, portal vein, pulmonary artery, and ureters. Using high-resolution CT segmentations of the liver, lungs, kidneys, and bladder, we incorporate organ-specific blood supply sources to improve kinetic modelling. Our method was evaluated on dynamic [$^{18}$F]FDG PET data from nine patients, resulting in a mean squared error (MSE) reduction of $13.39\%$ for the liver and $10.42\%$ for the lungs. These initial results highlight the potential of multiple IDIFs in improving anatomical modelling and fully leveraging dynamic PET imaging. This approach could facilitate the integration of tracer kinetic modelling into clinical routine.
Fine-Tuning TransMorph with Gradient Correlation for Anatomical Alignment
Dec 30, 2024



Abstract:Unsupervised deep learning is a promising method in brain MRI registration to reduce the reliance on anatomical labels, while still achieving anatomically accurate transformations. For the Learn2Reg2024 LUMIR challenge, we propose fine-tuning of the pre-trained TransMorph model to improve the convergence stability as well as the deformation smoothness. The former is achieved through the FAdam optimizer, and consistency in structural changes is incorporated through the addition of gradient correlation in the similarity measure, improving anatomical alignment. The results show slight improvements in the Dice and HdDist95 scores, and a notable reduction in the NDV compared to the baseline TransMorph model. These are also confirmed by inspecting the boundaries of the tissue. Our proposed method highlights the effectiveness of including Gradient Correlation to achieve smoother and structurally consistent deformations for interpatient brain MRI registration.
Shape Completion in the Dark: Completing Vertebrae Morphology from 3D Ultrasound
Apr 11, 2024Abstract:Purpose: Ultrasound (US) imaging, while advantageous for its radiation-free nature, is challenging to interpret due to only partially visible organs and a lack of complete 3D information. While performing US-based diagnosis or investigation, medical professionals therefore create a mental map of the 3D anatomy. In this work, we aim to replicate this process and enhance the visual representation of anatomical structures. Methods: We introduce a point-cloud-based probabilistic DL method to complete occluded anatomical structures through 3D shape completion and choose US-based spine examinations as our application. To enable training, we generate synthetic 3D representations of partially occluded spinal views by mimicking US physics and accounting for inherent artifacts. Results: The proposed model performs consistently on synthetic and patient data, with mean and median differences of 2.02 and 0.03 in CD, respectively. Our ablation study demonstrates the importance of US physics-based data generation, reflected in the large mean and median difference of 11.8 CD and 9.55 CD, respectively. Additionally, we demonstrate that anatomic landmarks, such as the spinous process (with reconstruction CD of 4.73) and the facet joints (mean distance to GT of 4.96mm) are preserved in the 3D completion. Conclusion: Our work establishes the feasibility of 3D shape completion for lumbar vertebrae, ensuring the preservation of level-wise characteristics and successful generalization from synthetic to real data. The incorporation of US physics contributes to more accurate patient data completions. Notably, our method preserves essential anatomic landmarks and reconstructs crucial injections sites at their correct locations. The generated data and source code will be made publicly available (https://github.com/miruna20/Shape-Completion-in-the-Dark).
AutoPaint: A Self-Inpainting Method for Unsupervised Anomaly Detection
May 21, 2023Abstract:Robust and accurate detection and segmentation of heterogenous tumors appearing in different anatomical organs with supervised methods require large-scale labeled datasets covering all possible types of diseases. Due to the unavailability of such rich datasets and the high cost of annotations, unsupervised anomaly detection (UAD) methods have been developed aiming to detect the pathologies as deviation from the normality by utilizing the unlabeled healthy image data. However, developed UAD models are often trained with an incomplete distribution of healthy anatomies and have difficulties in preserving anatomical constraints. This work intends to, first, propose a robust inpainting model to learn the details of healthy anatomies and reconstruct high-resolution images by preserving anatomical constraints. Second, we propose an autoinpainting pipeline to automatically detect tumors, replace their appearance with the learned healthy anatomies, and based on that segment the tumoral volumes in a purely unsupervised fashion. Three imaging datasets, including PET, CT, and PET-CT scans of lung tumors and head and neck tumors, are studied as benchmarks for evaluation. Experimental results demonstrate the significant superiority of the proposed method over a wide range of state-of-the-art UAD methods. Moreover, the unsupervised method we propose produces comparable results to a robust supervised segmentation method when applied to multimodal images.
Self-Supervised Learning for Physiologically-Based Pharmacokinetic Modeling in Dynamic PET
May 17, 2023



Abstract:Dynamic positron emission tomography imaging (dPET) provides temporally resolved images of a tracer enabling a quantitative measure of physiological processes. Voxel-wise physiologically-based pharmacokinetic (PBPK) modeling of the time activity curves (TAC) can provide relevant diagnostic information for clinical workflow. Conventional fitting strategies for TACs are slow and ignore the spatial relation between neighboring voxels. We train a spatio-temporal UNet to estimate the kinetic parameters given TAC from F-18-fluorodeoxyglucose (FDG) dPET. This work introduces a self-supervised loss formulation to enforce the similarity between the measured TAC and those generated with the learned kinetic parameters. Our method provides quantitatively comparable results at organ-level to the significantly slower conventional approaches, while generating pixel-wise parametric images which are consistent with expected physiology. To the best of our knowledge, this is the first self-supervised network that allows voxel-wise computation of kinetic parameters consistent with a non-linear kinetic model. The code will become publicly available upon acceptance.
Precise Repositioning of Robotic Ultrasound: Improving Registration-based Motion Compensation using Ultrasound Confidence Optimization
Aug 10, 2022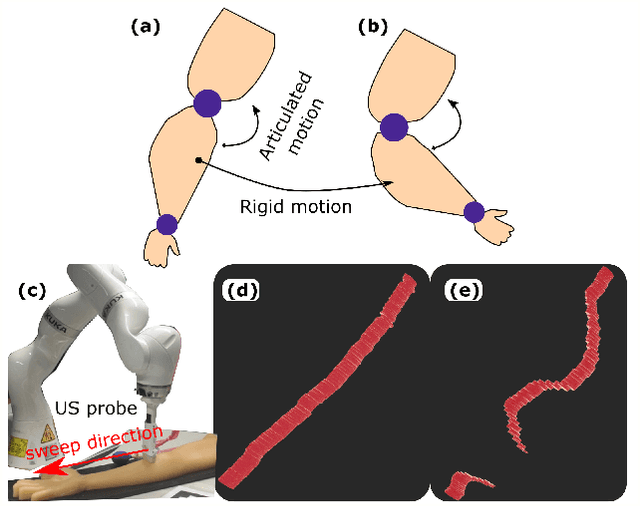

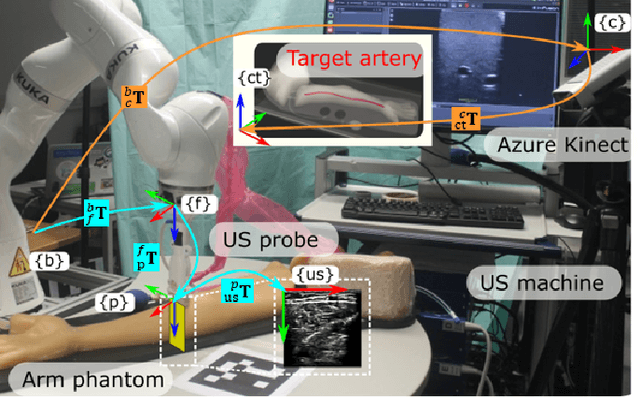

Abstract:Robotic ultrasound (US) imaging has been seen as a promising solution to overcome the limitations of free-hand US examinations, i.e., inter-operator variability. \revision{However, the fact that robotic US systems cannot react to subject movements during scans limits their clinical acceptance.} Regarding human sonographers, they often react to patient movements by repositioning the probe or even restarting the acquisition, in particular for the scans of anatomies with long structures like limb arteries. To realize this characteristic, we proposed a vision-based system to monitor the subject's movement and automatically update the scan trajectory thus seamlessly obtaining a complete 3D image of the target anatomy. The motion monitoring module is developed using the segmented object masks from RGB images. Once the subject is moved, the robot will stop and recompute a suitable trajectory by registering the surface point clouds of the object obtained before and after the movement using the iterative closest point algorithm. Afterward, to ensure optimal contact conditions after repositioning US probe, a confidence-based fine-tuning process is used to avoid potential gaps between the probe and contact surface. Finally, the whole system is validated on a human-like arm phantom with an uneven surface, while the object segmentation network is also validated on volunteers. The results demonstrate that the presented system can react to object movements and reliably provide accurate 3D images.
U-PET: MRI-based Dementia Detection with Joint Generation of Synthetic FDG-PET Images
Jun 16, 2022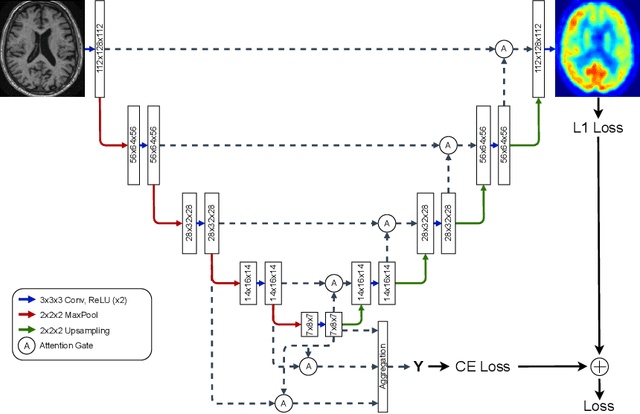
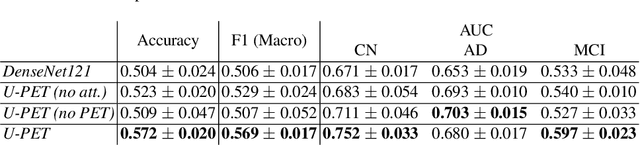
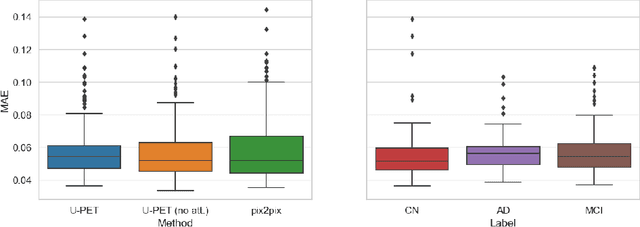
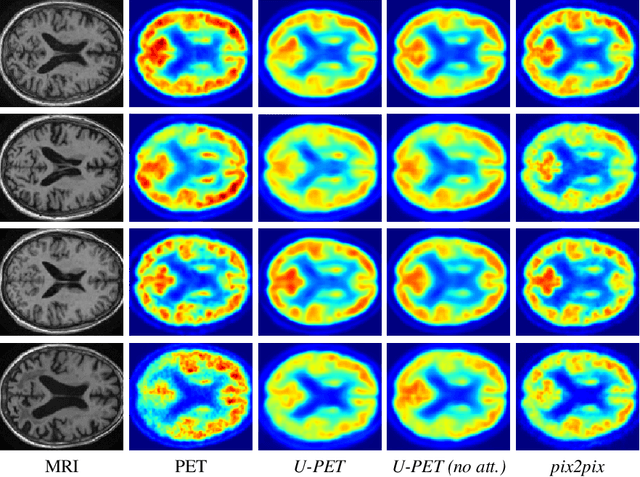
Abstract:Alzheimer's disease (AD) is the most common cause of dementia. An early detection is crucial for slowing down the disease and mitigating risks related to the progression. While the combination of MRI and FDG-PET is the best image-based tool for diagnosis, FDG-PET is not always available. The reliable detection of Alzheimer's disease with only MRI could be beneficial, especially in regions where FDG-PET might not be affordable for all patients. To this end, we propose a multi-task method based on U-Net that takes T1-weighted MR images as an input to generate synthetic FDG-PET images and classifies the dementia progression of the patient into cognitive normal (CN), cognitive impairment (MCI), and AD. The attention gates used in both task heads can visualize the most relevant parts of the brain, guiding the examiner and adding interpretability. Results show the successful generation of synthetic FDG-PET images and a performance increase in disease classification over the naive single-task baseline.
Weakly-supervised Biomechanically-constrained CT/MRI Registration of the Spine
May 16, 2022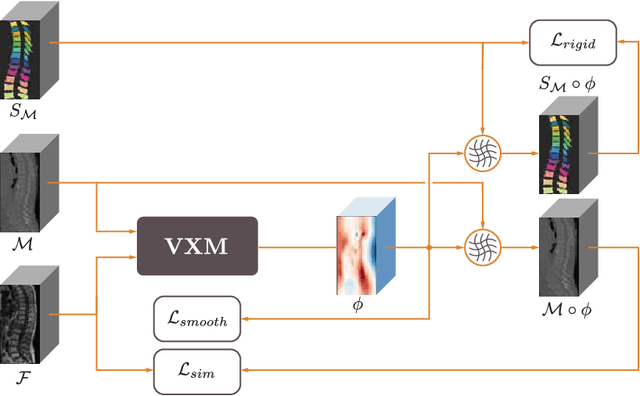
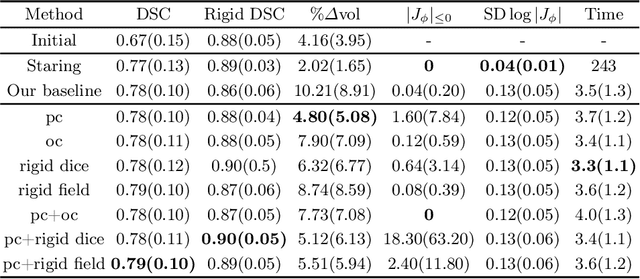
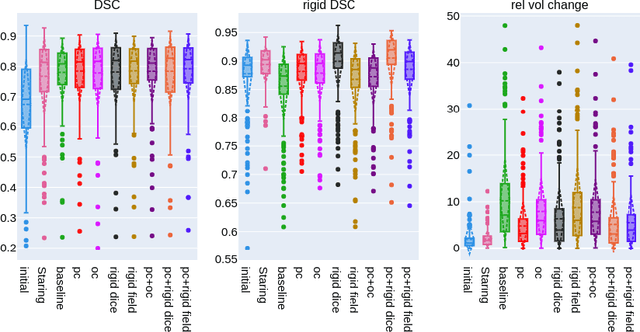
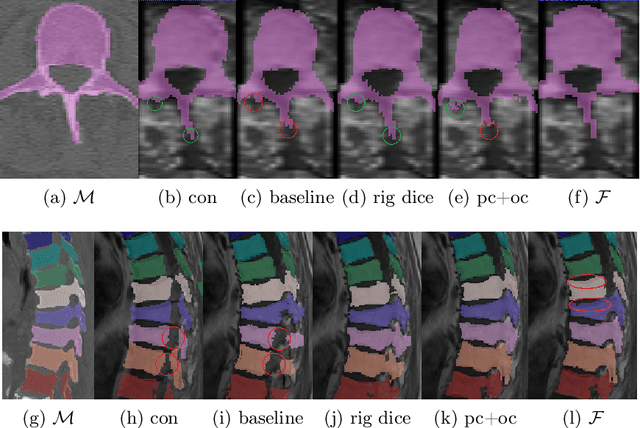
Abstract:CT and MRI are two of the most informative modalities in spinal diagnostics and treatment planning. CT is useful when analysing bony structures, while MRI gives information about the soft tissue. Thus, fusing the information of both modalities can be very beneficial. Registration is the first step for this fusion. While the soft tissues around the vertebra are deformable, each vertebral body is constrained to move rigidly. We propose a weakly-supervised deep learning framework that preserves the rigidity and the volume of each vertebra while maximizing the accuracy of the registration. To achieve this goal, we introduce anatomy-aware losses for training the network. We specifically design these losses to depend only on the CT label maps since automatic vertebra segmentation in CT gives more accurate results contrary to MRI. We evaluate our method on an in-house dataset of 167 patients. Our results show that adding the anatomy-aware losses increases the plausibility of the inferred transformation while keeping the accuracy untouched.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge