Örjan Smedby
Anatomy-Aware Lymphoma Lesion Detection in Whole-Body PET/CT
Nov 10, 2025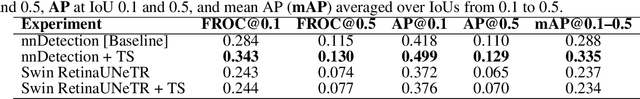
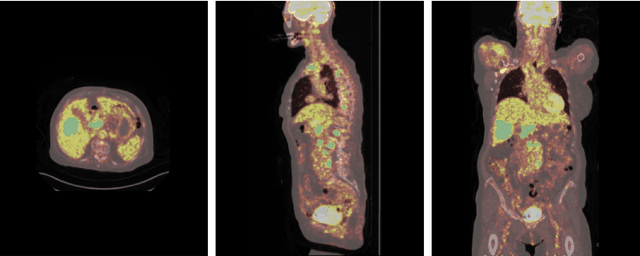
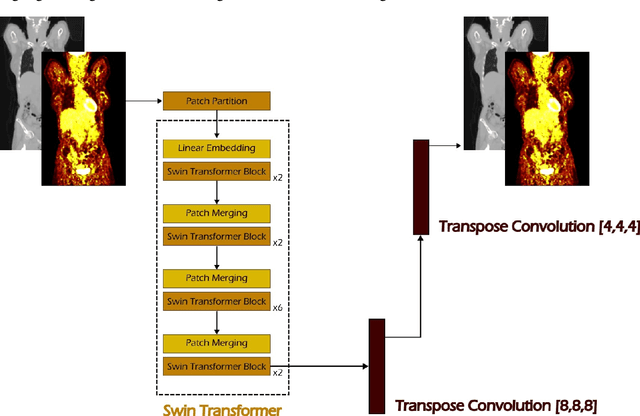
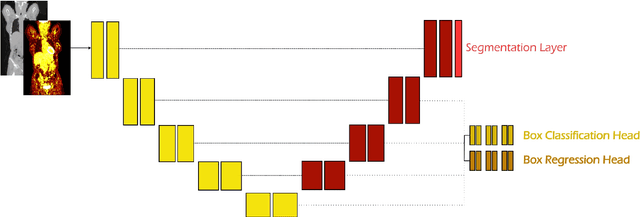
Abstract:Early cancer detection is crucial for improving patient outcomes, and 18F FDG PET/CT imaging plays a vital role by combining metabolic and anatomical information. Accurate lesion detection remains challenging due to the need to identify multiple lesions of varying sizes. In this study, we investigate the effect of adding anatomy prior information to deep learning-based lesion detection models. In particular, we add organ segmentation masks from the TotalSegmentator tool as auxiliary inputs to provide anatomical context to nnDetection, which is the state-of-the-art for lesion detection, and Swin Transformer. The latter is trained in two stages that combine self-supervised pre-training and supervised fine-tuning. The method is tested in the AutoPET and Karolinska lymphoma datasets. The results indicate that the inclusion of anatomical priors substantially improves the detection performance within the nnDetection framework, while it has almost no impact on the performance of the vision transformer. Moreover, we observe that Swin Transformer does not offer clear advantages over conventional convolutional neural network (CNN) encoders used in nnDetection. These findings highlight the critical role of the anatomical context in cancer lesion detection, especially in CNN-based models.
Unsupervised Domain Adaptation for Pediatric Brain Tumor Segmentation
Jun 24, 2024



Abstract:Significant advances have been made toward building accurate automatic segmentation models for adult gliomas. However, the performance of these models often degrades when applied to pediatric glioma due to their imaging and clinical differences (domain shift). Obtaining sufficient annotated data for pediatric glioma is typically difficult because of its rare nature. Also, manual annotations are scarce and expensive. In this work, we propose Domain-Adapted nnU-Net (DA-nnUNet) to perform unsupervised domain adaptation from adult glioma (source domain) to pediatric glioma (target domain). Specifically, we add a domain classifier connected with a gradient reversal layer (GRL) to a backbone nnU-Net. Once the classifier reaches a very high accuracy, the GRL is activated with the goal of transferring domain-invariant features from the classifier to the segmentation model while preserving segmentation accuracy on the source domain. The accuracy of the classifier slowly degrades to chance levels. No annotations are used in the target domain. The method is compared to 8 different supervised models using BraTS-Adult glioma (N=1251) and BraTS-PED glioma data (N=99). The proposed method shows notable performance enhancements in the tumor core (TC) region compared to the model that only uses adult data: ~32% better Dice scores and ~20 better 95th percentile Hausdorff distances. Moreover, our unsupervised approach shows no statistically significant difference compared to the practical upper bound model using manual annotations from both datasets in TC region. The code is shared at https://github.com/Fjr9516/DA_nnUNet.
A deformation-based morphometry framework for disentangling Alzheimer's disease from normal aging using learned normal aging templates
Nov 14, 2023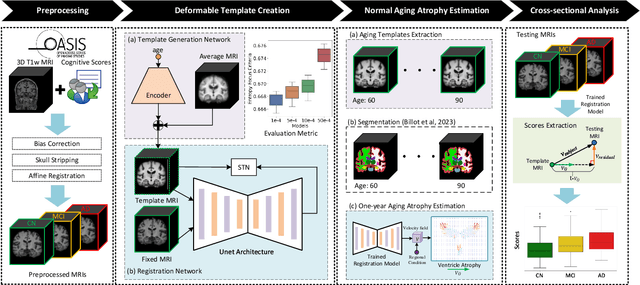
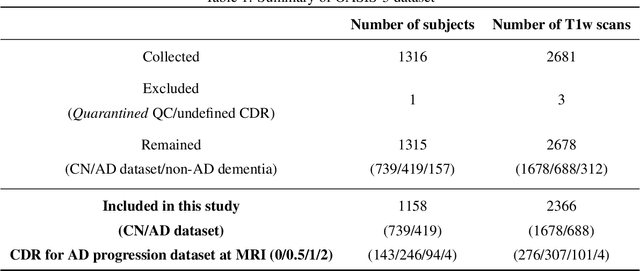
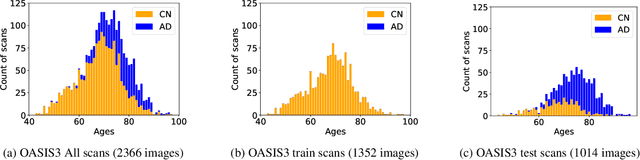
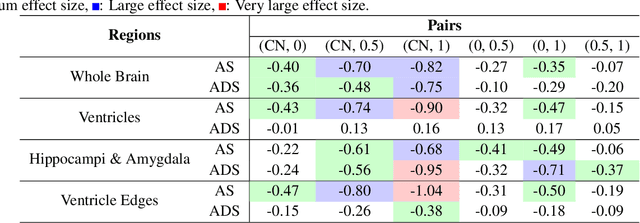
Abstract:Alzheimer's Disease and normal aging are both characterized by brain atrophy. The question of whether AD-related brain atrophy represents accelerated aging or a neurodegeneration process distinct from that in normal aging remains unresolved. Moreover, precisely disentangling AD-related brain atrophy from normal aging in a clinical context is complex. In this study, we propose a deformation-based morphometry framework to estimate normal aging and AD-specific atrophy patterns of subjects from morphological MRI scans. We first leverage deep-learning-based methods to create age-dependent templates of cognitively normal (CN) subjects. These templates model the normal aging atrophy patterns in a CN population. Then, we use the learned diffeomorphic registration to estimate the one-year normal aging pattern at the voxel level. We register the testing image to the 60-year-old CN template in the second step. Finally, normal aging and AD-specific scores are estimated by measuring the alignment of this registration with the one-year normal aging pattern. The methodology was developed and evaluated on the OASIS3 dataset with 1,014 T1-weighted MRI scans. Of these, 326 scans were from CN subjects, and 688 scans were from individuals clinically diagnosed with AD at different stages of clinical severity defined by clinical dementia rating (CDR) scores. The results show that ventricles predominantly follow an accelerated normal aging pattern in subjects with AD. In turn, hippocampi and amygdala regions were affected by both normal aging and AD-specific factors. Interestingly, hippocampi and amygdala regions showed more of an accelerated normal aging pattern for subjects during the early clinical stages of the disease, while the AD-specific score increases in later clinical stages. Our code is freely available at https://github.com/Fjr9516/DBM_with_DL.
AutoPaint: A Self-Inpainting Method for Unsupervised Anomaly Detection
May 21, 2023Abstract:Robust and accurate detection and segmentation of heterogenous tumors appearing in different anatomical organs with supervised methods require large-scale labeled datasets covering all possible types of diseases. Due to the unavailability of such rich datasets and the high cost of annotations, unsupervised anomaly detection (UAD) methods have been developed aiming to detect the pathologies as deviation from the normality by utilizing the unlabeled healthy image data. However, developed UAD models are often trained with an incomplete distribution of healthy anatomies and have difficulties in preserving anatomical constraints. This work intends to, first, propose a robust inpainting model to learn the details of healthy anatomies and reconstruct high-resolution images by preserving anatomical constraints. Second, we propose an autoinpainting pipeline to automatically detect tumors, replace their appearance with the learned healthy anatomies, and based on that segment the tumoral volumes in a purely unsupervised fashion. Three imaging datasets, including PET, CT, and PET-CT scans of lung tumors and head and neck tumors, are studied as benchmarks for evaluation. Experimental results demonstrate the significant superiority of the proposed method over a wide range of state-of-the-art UAD methods. Moreover, the unsupervised method we propose produces comparable results to a robust supervised segmentation method when applied to multimodal images.
Development and evaluation of a 3D annotation software for interactive COVID-19 lesion segmentation in chest CT
Jan 13, 2021



Abstract:Segmentation of COVID-19 lesions from chest CT scans is of great importance for better diagnosing the disease and investigating its extent. However, manual segmentation can be very time consuming and subjective, given the lesions' large variation in shape, size and position. On the other hand, we still lack large manually segmented datasets that could be used for training machine learning-based models for fully automatic segmentation. In this work, we propose a new interactive and user-friendly tool for COVID-19 lesion segmentation, which works by alternating automatic steps (based on level-set segmentation and statistical shape modeling) with manual correction steps. The present software was tested by two different expertise groups: one group of three radiologists and one of three users with an engineering background. Promising segmentation results were obtained by both groups, which achieved satisfactory agreement both between- and within-group. Moreover, our interactive tool was shown to significantly speed up the lesion segmentation process, when compared to fully manual segmentation. Finally, we investigated inter-observer variability and how it is strongly influenced by several subjective factors, showing the importance for AI researchers and clinical doctors to be aware of the uncertainty in lesion segmentation results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge