Chunliang Wang
AutoPaint: A Self-Inpainting Method for Unsupervised Anomaly Detection
May 21, 2023Abstract:Robust and accurate detection and segmentation of heterogenous tumors appearing in different anatomical organs with supervised methods require large-scale labeled datasets covering all possible types of diseases. Due to the unavailability of such rich datasets and the high cost of annotations, unsupervised anomaly detection (UAD) methods have been developed aiming to detect the pathologies as deviation from the normality by utilizing the unlabeled healthy image data. However, developed UAD models are often trained with an incomplete distribution of healthy anatomies and have difficulties in preserving anatomical constraints. This work intends to, first, propose a robust inpainting model to learn the details of healthy anatomies and reconstruct high-resolution images by preserving anatomical constraints. Second, we propose an autoinpainting pipeline to automatically detect tumors, replace their appearance with the learned healthy anatomies, and based on that segment the tumoral volumes in a purely unsupervised fashion. Three imaging datasets, including PET, CT, and PET-CT scans of lung tumors and head and neck tumors, are studied as benchmarks for evaluation. Experimental results demonstrate the significant superiority of the proposed method over a wide range of state-of-the-art UAD methods. Moreover, the unsupervised method we propose produces comparable results to a robust supervised segmentation method when applied to multimodal images.
Development and evaluation of a 3D annotation software for interactive COVID-19 lesion segmentation in chest CT
Jan 13, 2021



Abstract:Segmentation of COVID-19 lesions from chest CT scans is of great importance for better diagnosing the disease and investigating its extent. However, manual segmentation can be very time consuming and subjective, given the lesions' large variation in shape, size and position. On the other hand, we still lack large manually segmented datasets that could be used for training machine learning-based models for fully automatic segmentation. In this work, we propose a new interactive and user-friendly tool for COVID-19 lesion segmentation, which works by alternating automatic steps (based on level-set segmentation and statistical shape modeling) with manual correction steps. The present software was tested by two different expertise groups: one group of three radiologists and one of three users with an engineering background. Promising segmentation results were obtained by both groups, which achieved satisfactory agreement both between- and within-group. Moreover, our interactive tool was shown to significantly speed up the lesion segmentation process, when compared to fully manual segmentation. Finally, we investigated inter-observer variability and how it is strongly influenced by several subjective factors, showing the importance for AI researchers and clinical doctors to be aware of the uncertainty in lesion segmentation results.
Investigating and Exploiting Image Resolution for Transfer Learning-based Skin Lesion Classification
Jun 25, 2020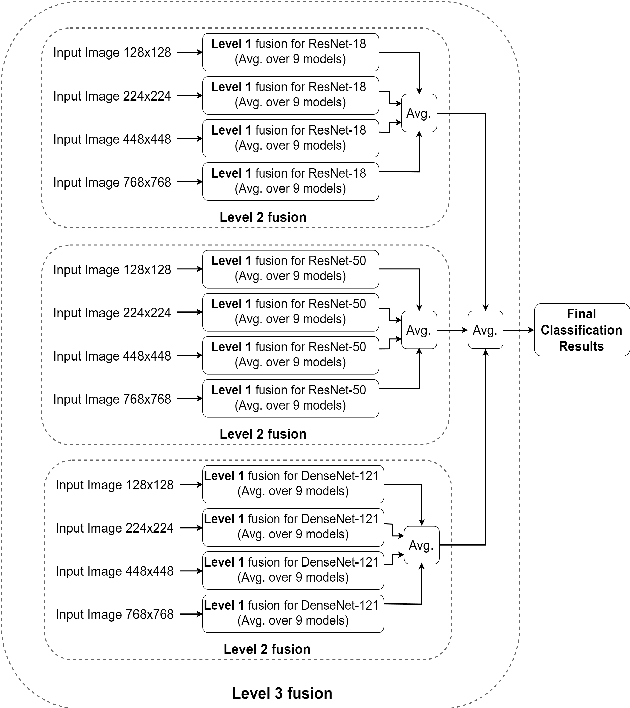

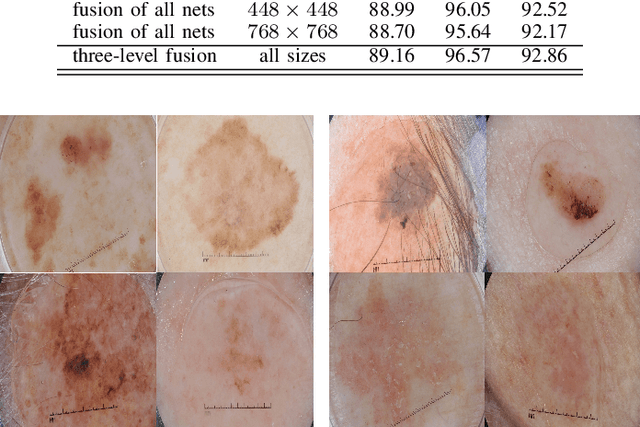

Abstract:Skin cancer is among the most common cancer types. Dermoscopic image analysis improves the diagnostic accuracy for detection of malignant melanoma and other pigmented skin lesions when compared to unaided visual inspection. Hence, computer-based methods to support medical experts in the diagnostic procedure are of great interest. Fine-tuning pre-trained convolutional neural networks (CNNs) has been shown to work well for skin lesion classification. Pre-trained CNNs are usually trained with natural images of a fixed image size which is typically significantly smaller than captured skin lesion images and consequently dermoscopic images are downsampled for fine-tuning. However, useful medical information may be lost during this transformation. In this paper, we explore the effect of input image size on skin lesion classification performance of fine-tuned CNNs. For this, we resize dermoscopic images to different resolutions, ranging from 64x64 to 768x768 pixels and investigate the resulting classification performance of three well-established CNNs, namely DenseNet-121, ResNet-18, and ResNet-50. Our results show that using very small images (of size 64x64 pixels) degrades the classification performance, while images of size 128x128 pixels and above support good performance with larger image sizes leading to slightly improved classification. We further propose a novel fusion approach based on a three-level ensemble strategy that exploits multiple fine-tuned networks trained with dermoscopic images at various sizes. When applied on the ISIC 2017 skin lesion classification challenge, our fusion approach yields an area under the receiver operating characteristic curve of 89.2% and 96.6% for melanoma classification and seborrheic keratosis classification, respectively, outperforming state-of-the-art algorithms.
Evaluation of Algorithms for Multi-Modality Whole Heart Segmentation: An Open-Access Grand Challenge
Feb 21, 2019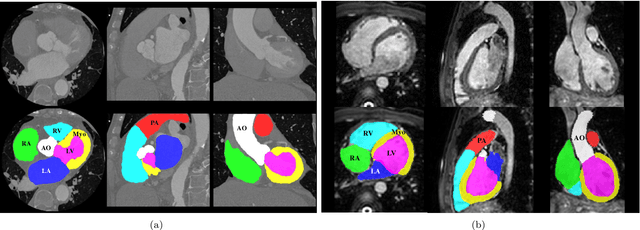
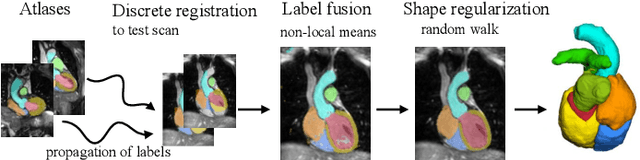
Abstract:Knowledge of whole heart anatomy is a prerequisite for many clinical applications. Whole heart segmentation (WHS), which delineates substructures of the heart, can be very valuable for modeling and analysis of the anatomy and functions of the heart. However, automating this segmentation can be arduous due to the large variation of the heart shape, and different image qualities of the clinical data. To achieve this goal, a set of training data is generally needed for constructing priors or for training. In addition, it is difficult to perform comparisons between different methods, largely due to differences in the datasets and evaluation metrics used. This manuscript presents the methodologies and evaluation results for the WHS algorithms selected from the submissions to the Multi-Modality Whole Heart Segmentation (MM-WHS) challenge, in conjunction with MICCAI 2017. The challenge provides 120 three-dimensional cardiac images covering the whole heart, including 60 CT and 60 MRI volumes, all acquired in clinical environments with manual delineation. Ten algorithms for CT data and eleven algorithms for MRI data, submitted from twelve groups, have been evaluated. The results show that many of the deep learning (DL) based methods achieved high accuracy, even though the number of training datasets was limited. A number of them also reported poor results in the blinded evaluation, probably due to overfitting in their training. The conventional algorithms, mainly based on multi-atlas segmentation, demonstrated robust and stable performance, even though the accuracy is not as good as the best DL method in CT segmentation. The challenge, including the provision of the annotated training data and the blinded evaluation for submitted algorithms on the test data, continues as an ongoing benchmarking resource via its homepage (\url{www.sdspeople.fudan.edu.cn/zhuangxiahai/0/mmwhs/}).
AVRA: Automatic Visual Ratings of Atrophy from MRI images using Recurrent Convolutional Neural Networks
Dec 23, 2018



Abstract:Quantifying the degree of atrophy is done clinically by neuroradiologists following established visual rating scales. For these assessments to be reliable the rater requires substantial training and experience, and even then the rating agreement between two radiologists is not perfect. We have developed a model we call AVRA (Automatic Visual Ratings of Atrophy) based on machine learning methods and trained on 2350 visual ratings made by an experienced neuroradiologist. It provides fast and automatic ratings for Scheltens' scale of medial temporal atrophy (MTA), the frontal subscale of Pasquier's Global Cortical Atrophy (GCA-F) scale, and Koedam's scale of Posterior Atrophy (PA). We demonstrate substantial inter-rater agreement between AVRA's and a neuroradiologist ratings with Cohen's weighted kappa values of $\kappa_w$ = 0.74/0.72 (MTA left/right), $\kappa_w$ = 0.62 (GCA-F) and $\kappa_w$ = 0.74 (PA), with an inherent intra-rater agreement of $\kappa_w$ = 1. We conclude that automatic visual ratings of atrophy can potentially have great clinical and scientific value, and aim to present AVRA as a freely available toolbox.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge