Georg Dorffner
Improving Generalization Capability of Deep Learning-Based Nuclei Instance Segmentation by Non-deterministic Train Time and Deterministic Test Time Stain Normalization
Sep 12, 2023



Abstract:With the advent of digital pathology and microscopic systems that can scan and save whole slide histological images automatically, there is a growing trend to use computerized methods to analyze acquired images. Among different histopathological image analysis tasks, nuclei instance segmentation plays a fundamental role in a wide range of clinical and research applications. While many semi- and fully-automatic computerized methods have been proposed for nuclei instance segmentation, deep learning (DL)-based approaches have been shown to deliver the best performances. However, the performance of such approaches usually degrades when tested on unseen datasets. In this work, we propose a novel approach to improve the generalization capability of a DL-based automatic segmentation approach. Besides utilizing one of the state-of-the-art DL-based models as a baseline, our method incorporates non-deterministic train time and deterministic test time stain normalization. We trained the model with one single training set and evaluated its segmentation performance on seven test datasets. Our results show that the proposed method provides up to 5.77%, 5.36%, and 5.27% better performance in segmenting nuclei based on Dice score, aggregated Jaccard index, and panoptic quality score, respectively, compared to the baseline segmentation model.
NuInsSeg: A Fully Annotated Dataset for Nuclei Instance Segmentation in H&E-Stained Histological Images
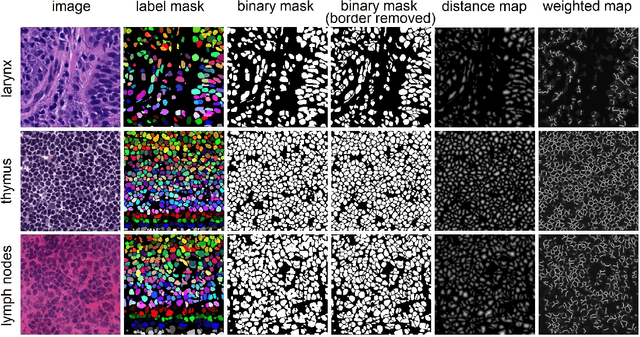
Aug 03, 2023Abstract:In computational pathology, automatic nuclei instance segmentation plays an essential role in whole slide image analysis. While many computerized approaches have been proposed for this task, supervised deep learning (DL) methods have shown superior segmentation performances compared to classical machine learning and image processing techniques. However, these models need fully annotated datasets for training which is challenging to acquire, especially in the medical domain. In this work, we release one of the biggest fully manually annotated datasets of nuclei in Hematoxylin and Eosin (H&E)-stained histological images, called NuInsSeg. This dataset contains 665 image patches with more than 30,000 manually segmented nuclei from 31 human and mouse organs. Moreover, for the first time, we provide additional ambiguous area masks for the entire dataset. These vague areas represent the parts of the images where precise and deterministic manual annotations are impossible, even for human experts. The dataset and detailed step-by-step instructions to generate related segmentation masks are publicly available at https://www.kaggle.com/datasets/ipateam/nuinsseg and https://github.com/masih4/NuInsSeg, respectively.
A global analysis of metrics used for measuring performance in natural language processing
Apr 25, 2022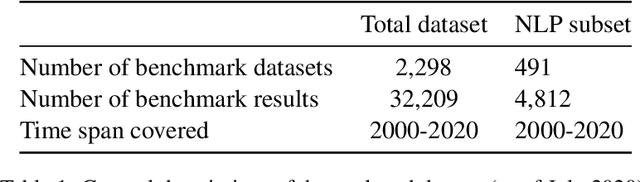
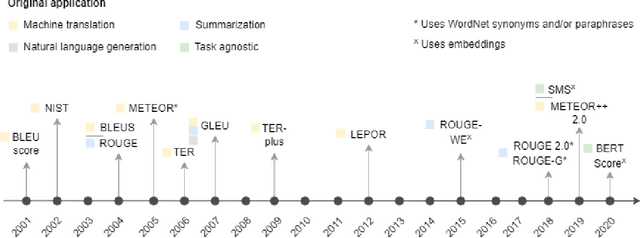
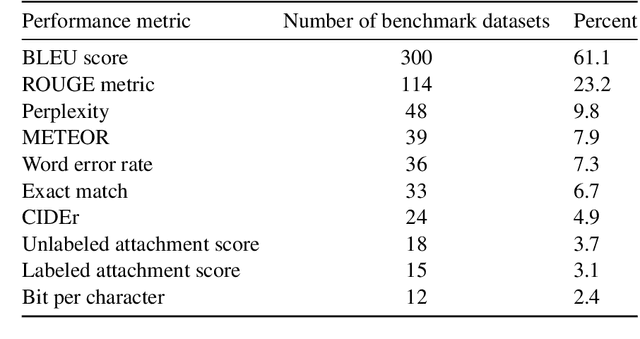
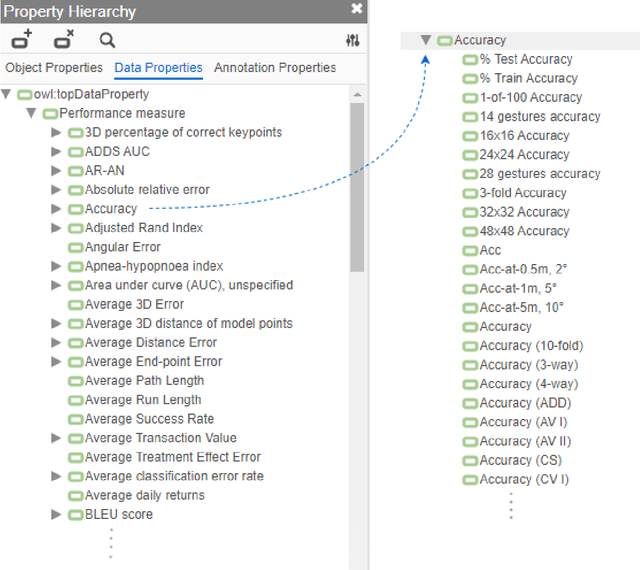
Abstract:Measuring the performance of natural language processing models is challenging. Traditionally used metrics, such as BLEU and ROUGE, originally devised for machine translation and summarization, have been shown to suffer from low correlation with human judgment and a lack of transferability to other tasks and languages. In the past 15 years, a wide range of alternative metrics have been proposed. However, it is unclear to what extent this has had an impact on NLP benchmarking efforts. Here we provide the first large-scale cross-sectional analysis of metrics used for measuring performance in natural language processing. We curated, mapped and systematized more than 3500 machine learning model performance results from the open repository 'Papers with Code' to enable a global and comprehensive analysis. Our results suggest that the large majority of natural language processing metrics currently used have properties that may result in an inadequate reflection of a models' performance. Furthermore, we found that ambiguities and inconsistencies in the reporting of metrics may lead to difficulties in interpreting and comparing model performances, impairing transparency and reproducibility in NLP research.
CryoNuSeg: A Dataset for Nuclei Instance Segmentation of Cryosectioned H&E-Stained Histological Images
Jan 02, 2021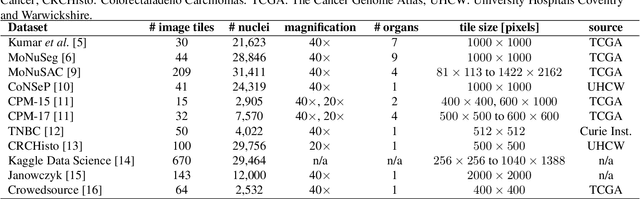
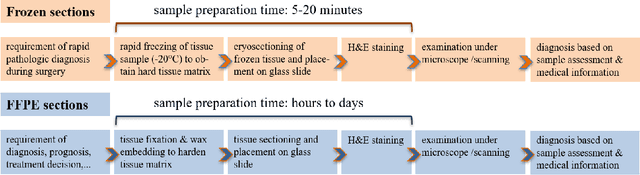
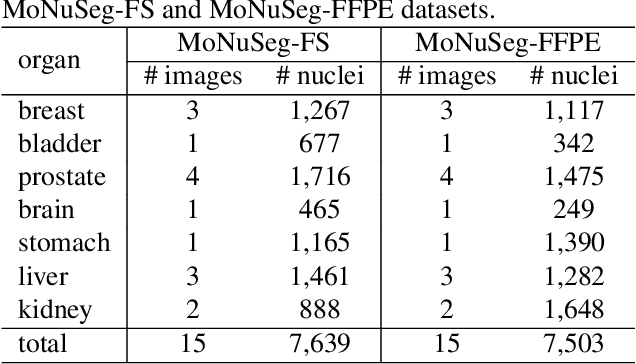
Abstract:Nuclei instance segmentation plays an important role in the analysis of Hematoxylin and Eosin (H&E)-stained images. While supervised deep learning (DL)-based approaches represent the state-of-the-art in automatic nuclei instance segmentation, annotated datasets are required to train these models. There are two main types of tissue processing protocols, namely formalin-fixed paraffin-embedded samples (FFPE) and frozen tissue samples (FS). Although FFPE-derived H&E stained tissue sections are the most widely used samples, H&E staining on frozen sections derived from FS samples is a relevant method in intra-operative surgical sessions as it can be performed fast. Due to differences in the protocols of these two types of samples, the derived images and in particular the nuclei appearance may be different in the acquired whole slide images. Analysis of FS-derived H&E stained images can be more challenging as rapid preparation, staining, and scanning of FS sections may lead to deterioration in image quality. In this paper, we introduce CryoNuSeg, the first fully annotated FS-derived cryosectioned and H&E-stained nuclei instance segmentation dataset. The dataset contains images from 10 human organs that were not exploited in other publicly available datasets, and is provided with three manual mark-ups to allow measuring intra-observer and inter-observer variability. Moreover, we investigate the effects of tissue fixation/embedding protocol (i.e., FS or FFPE) on the automatic nuclei instance segmentation performance of one of the state-of-the-art DL approaches. We also create a baseline segmentation benchmark for the dataset that can be used in future research. A step-by-step guide to generate the dataset as well as the full dataset and other detailed information are made available to fellow researchers at https://github.com/masih4/CryoNuSeg.
A critical analysis of metrics used for measuring progress in artificial intelligence
Aug 06, 2020



Abstract:Comparing model performances on benchmark datasets is an integral part of measuring and driving progress in artificial intelligence. A model's performance on a benchmark dataset is commonly assessed based on a single or a small set of performance metrics. While this enables quick comparisons, it may also entail the risk of inadequately reflecting model performance if the metric does not sufficiently cover all performance characteristics. Currently, it is unknown to what extent this might impact current benchmarking efforts. To address this question, we analysed the current landscape of performance metrics based on data covering 3867 machine learning model performance results from the web-based open platform 'Papers with Code'. Our results suggest that the large majority of metrics currently used to evaluate classification AI benchmark tasks have properties that may result in an inadequate reflection of a classifiers' performance, especially when used with imbalanced datasets. While alternative metrics that address problematic properties have been proposed, they are currently rarely applied as performance metrics in benchmarking tasks. Finally, we noticed that the reporting of metrics was partly inconsistent and partly unspecific, which may lead to ambiguities when comparing model performances.
Investigating and Exploiting Image Resolution for Transfer Learning-based Skin Lesion Classification
Jun 25, 2020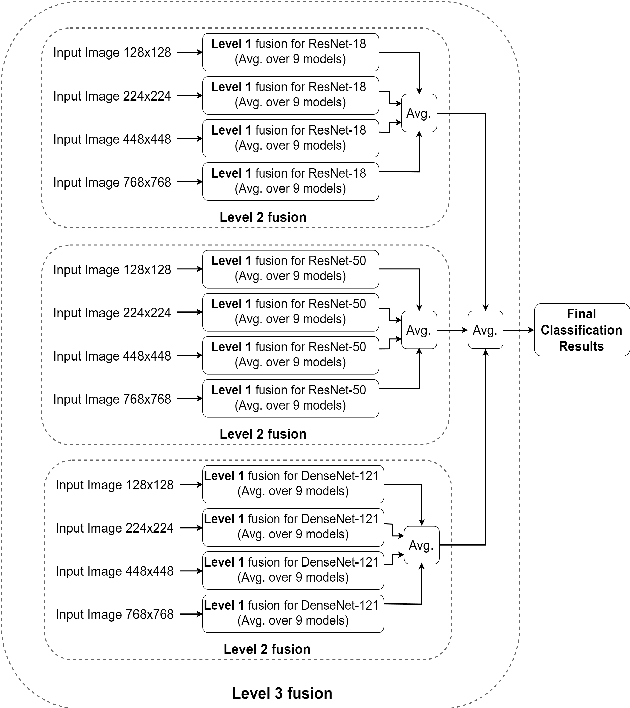

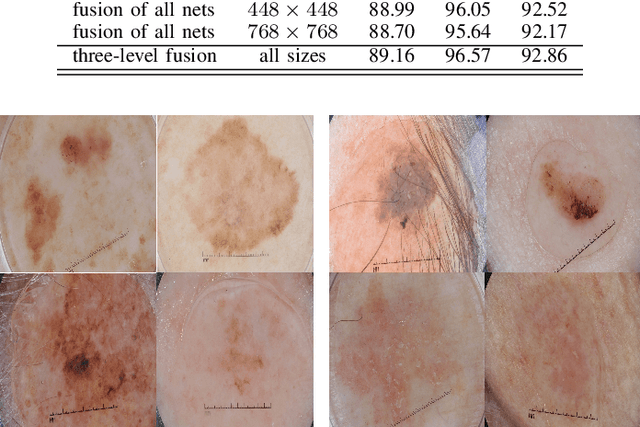

Abstract:Skin cancer is among the most common cancer types. Dermoscopic image analysis improves the diagnostic accuracy for detection of malignant melanoma and other pigmented skin lesions when compared to unaided visual inspection. Hence, computer-based methods to support medical experts in the diagnostic procedure are of great interest. Fine-tuning pre-trained convolutional neural networks (CNNs) has been shown to work well for skin lesion classification. Pre-trained CNNs are usually trained with natural images of a fixed image size which is typically significantly smaller than captured skin lesion images and consequently dermoscopic images are downsampled for fine-tuning. However, useful medical information may be lost during this transformation. In this paper, we explore the effect of input image size on skin lesion classification performance of fine-tuned CNNs. For this, we resize dermoscopic images to different resolutions, ranging from 64x64 to 768x768 pixels and investigate the resulting classification performance of three well-established CNNs, namely DenseNet-121, ResNet-18, and ResNet-50. Our results show that using very small images (of size 64x64 pixels) degrades the classification performance, while images of size 128x128 pixels and above support good performance with larger image sizes leading to slightly improved classification. We further propose a novel fusion approach based on a three-level ensemble strategy that exploits multiple fine-tuned networks trained with dermoscopic images at various sizes. When applied on the ISIC 2017 skin lesion classification challenge, our fusion approach yields an area under the receiver operating characteristic curve of 89.2% and 96.6% for melanoma classification and seborrheic keratosis classification, respectively, outperforming state-of-the-art algorithms.
Bayesian deep neural networks for low-cost neurophysiological markers of Alzheimer's disease severity
Dec 13, 2018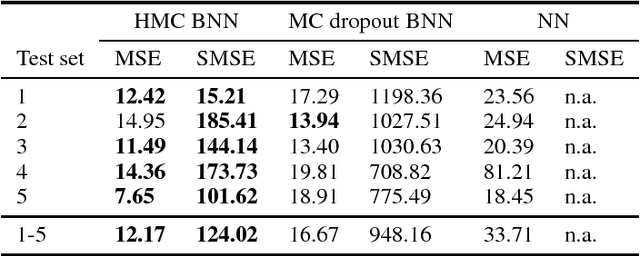
Abstract:As societies around the world are ageing, the number of Alzheimer's disease (AD) patients is rapidly increasing. To date, no low-cost, non-invasive biomarkers have been established to advance the objectivization of AD diagnosis and progression assessment. Here, we utilize Bayesian neural networks to develop a multivariate predictor for AD severity using a wide range of quantitative EEG (QEEG) markers. The Bayesian treatment of neural networks both automatically controls model complexity and provides a predictive distribution over the target function, giving uncertainty bounds for our regression task. It is therefore well suited to clinical neuroscience, where data sets are typically sparse and practitioners require a precise assessment of the predictive uncertainty. We use data of one of the largest prospective AD EEG trials ever conducted to demonstrate the potential of Bayesian deep learning in this domain, while comparing two distinct Bayesian neural network approaches, i.e., Monte Carlo dropout and Hamiltonian Monte Carlo.
Riemannian tangent space mapping and elastic net regularization for cost-effective EEG markers of brain atrophy in Alzheimer's disease
Nov 22, 2017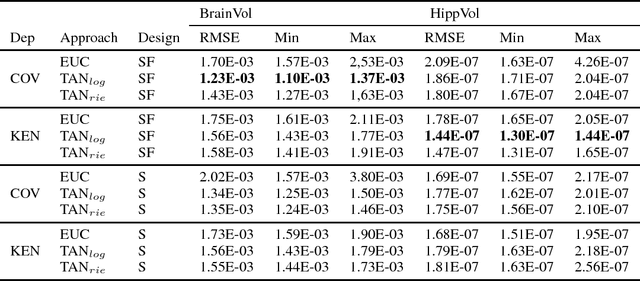
Abstract:The diagnosis of Alzheimer's disease (AD) in routine clinical practice is most commonly based on subjective clinical interpretations. Quantitative electroencephalography (QEEG) measures have been shown to reflect neurodegenerative processes in AD and might qualify as affordable and thereby widely available markers to facilitate the objectivization of AD assessment. Here, we present a novel framework combining Riemannian tangent space mapping and elastic net regression for the development of brain atrophy markers. While most AD QEEG studies are based on small sample sizes and psychological test scores as outcome measures, here we train and test our models using data of one of the largest prospective EEG AD trials ever conducted, including MRI biomarkers of brain atrophy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge