Ramona Woitek
NucFuseRank: Dataset Fusion and Performance Ranking for Nuclei Instance Segmentation
Jan 27, 2026Abstract:Nuclei instance segmentation in hematoxylin and eosin (H&E)-stained images plays an important role in automated histological image analysis, with various applications in downstream tasks. While several machine learning and deep learning approaches have been proposed for nuclei instance segmentation, most research in this field focuses on developing new segmentation algorithms and benchmarking them on a limited number of arbitrarily selected public datasets. In this work, rather than focusing on model development, we focused on the datasets used for this task. Based on an extensive literature review, we identified manually annotated, publicly available datasets of H&E-stained images for nuclei instance segmentation and standardized them into a unified input and annotation format. Using two state-of-the-art segmentation models, one based on convolutional neural networks (CNNs) and one based on a hybrid CNN and vision transformer architecture, we systematically evaluated and ranked these datasets based on their nuclei instance segmentation performance. Furthermore, we proposed a unified test set (NucFuse-test) for fair cross-dataset evaluation and a unified training set (NucFuse-train) for improved segmentation performance by merging images from multiple datasets. By evaluating and ranking the datasets, performing comprehensive analyses, generating fused datasets, conducting external validation, and making our implementation publicly available, we provided a new benchmark for training, testing, and evaluating nuclei instance segmentation models on H&E-stained histological images.
Developing Predictive and Robust Radiomics Models for Chemotherapy Response in High-Grade Serous Ovarian Carcinoma
Jan 13, 2026Abstract:Objectives: High-grade serous ovarian carcinoma (HGSOC) is typically diagnosed at an advanced stage with extensive peritoneal metastases, making treatment challenging. Neoadjuvant chemotherapy (NACT) is often used to reduce tumor burden before surgery, but about 40% of patients show limited response. Radiomics, combined with machine learning (ML), offers a promising non-invasive method for predicting NACT response by analyzing computed tomography (CT) imaging data. This study aimed to improve response prediction in HGSOC patients undergoing NACT by integration different feature selection methods. Materials and methods: A framework for selecting robust radiomics features was introduced by employing an automated randomisation algorithm to mimic inter-observer variability, ensuring a balance between feature robustness and prediction accuracy. Four response metrics were used: chemotherapy response score (CRS), RECIST, volume reduction (VolR), and diameter reduction (DiaR). Lesions in different anatomical sites were studied. Pre- and post-NACT CT scans were used for feature extraction and model training on one cohort, and an independent cohort was used for external testing. Results: The best prediction performance was achieved using all lesions combined for VolR prediction, with an AUC of 0.83. Omental lesions provided the best results for CRS prediction (AUC 0.77), while pelvic lesions performed best for DiaR (AUC 0.76). Conclusion: The integration of robustness into the feature selection processes ensures the development of reliable models and thus facilitates the implementation of the radiomics models in clinical applications for HGSOC patients. Future work should explore further applications of radiomics in ovarian cancer, particularly in real-time clinical settings.
ACS-SegNet: An Attention-Based CNN-SegFormer Segmentation Network for Tissue Segmentation in Histopathology
Oct 23, 2025Abstract:Automated histopathological image analysis plays a vital role in computer-aided diagnosis of various diseases. Among developed algorithms, deep learning-based approaches have demonstrated excellent performance in multiple tasks, including semantic tissue segmentation in histological images. In this study, we propose a novel approach based on attention-driven feature fusion of convolutional neural networks (CNNs) and vision transformers (ViTs) within a unified dual-encoder model to improve semantic segmentation performance. Evaluation on two publicly available datasets showed that our model achieved {\mu}IoU/{\mu}Dice scores of 76.79%/86.87% on the GCPS dataset and 64.93%/76.60% on the PUMA dataset, outperforming state-of-the-art and baseline benchmarks. The implementation of our method is publicly available in a GitHub repository: https://github.com/NimaTorbati/ACS-SegNet
Fusion of Foundation and Vision Transformer Model Features for Dermatoscopic Image Classification
May 22, 2025Abstract:Accurate classification of skin lesions from dermatoscopic images is essential for diagnosis and treatment of skin cancer. In this study, we investigate the utility of a dermatology-specific foundation model, PanDerm, in comparison with two Vision Transformer (ViT) architectures (ViT base and Swin Transformer V2 base) for the task of skin lesion classification. Using frozen features extracted from PanDerm, we apply non-linear probing with three different classifiers, namely, multi-layer perceptron (MLP), XGBoost, and TabNet. For the ViT-based models, we perform full fine-tuning to optimize classification performance. Our experiments on the HAM10000 and MSKCC datasets demonstrate that the PanDerm-based MLP model performs comparably to the fine-tuned Swin transformer model, while fusion of PanDerm and Swin Transformer predictions leads to further performance improvements. Future work will explore additional foundation models, fine-tuning strategies, and advanced fusion techniques.
Improved tissue sodium concentration quantification in breast cancer by reducing partial volume effects: a preliminary study
Mar 25, 2025



Abstract:Introduction: In sodium (23Na) MRI, partial volume effects (PVE) are one of the most common causes of errors in the quantification of tissue sodium concentration (TSC) in vivo. Advanced image reconstruction algorithms, such as compressed sensing (CS), have been shown to potentially reduce PVE. Therefore, we investigated the feasibility of CS-based methods for image quality and TSC quantification accuracy improvement in patients with breast cancer (BC). Subjects and Methods: Three healthy participants and 12 female participants with BC were examined on a 7T MRI scanner in this study. We reconstructed 23Na-MRI images using the weighted total variation (wTV) and directional total variation (dTV), anatomically guided total variation (AG-TV), and adaptive combine (ADC) reconstruction and performed image quality assessment. We evaluated agreement in tumor volumes delineated on sodium data using the Dice score and performed TSC quantification for different image reconstruction approaches. Results: All methods provided sodium images of the breast with good quality. The mean Dice scores for wTV, dTV, and AG-TV were 65%, 72%, and 75%, respectively. In the breast tumors, average TSC values were 83.0, 72.0, 80.0, and 84.0 mmol/L, respectively. There was a significant difference between dTV and wTV (p<0.001), as well as between dTV and AG-TV (p<0.001) and dTV and ADC algorithm (p<0.001). Conclusion: The results of this study showed that there are differences in tumor appearance and TSC estimations that might be depending on the type of image reconstruction and parameters used, most likely due to differences in their robustness in reducing PVE.
Variational U-Net with Local Alignment for Joint Tumor Extraction and Registration (VALOR-Net) of Breast MRI Data Acquired at Two Different Field Strengths
Jan 23, 2025



Abstract:Background: Multiparametric breast MRI data might improve tumor diagnostics, characterization, and treatment planning. Accurate alignment and delineation of images acquired at different field strengths such as 3T and 7T, remain challenging research tasks. Purpose: To address alignment challenges and enable consistent tumor segmentation across different MRI field strengths. Study type: Retrospective. Subjects: Nine female subjects with breast tumors were involved: six histologically proven invasive ductal carcinomas (IDC) and three fibroadenomas. Field strength/sequence: Imaging was performed at 3T and 7T scanners using post-contrast T1-weighted three-dimensional time-resolved angiography with stochastic trajectories (TWIST) sequence. Assessments: The method's performance for joint image registration and tumor segmentation was evaluated using several quantitative metrics, including signal-to-noise ratio (PSNR), structural similarity index (SSIM), normalized cross-correlation (NCC), Dice coefficient, F1 score, and relative sum of squared differences (rel SSD). Statistical tests: The Pearson correlation coefficient was used to test the relationship between the registration and segmentation metrics. Results: When calculated for each subject individually, the PSNR was in a range from 27.5 to 34.5 dB, and the SSIM was from 82.6 to 92.8%. The model achieved an NCC from 96.4 to 99.3% and a Dice coefficient of 62.9 to 95.3%. The F1 score was between 55.4 and 93.2% and the rel SSD was in the range of 2.0 and 7.5%. The segmentation metrics Dice and F1 Score are highly correlated (0.995), while a moderate correlation between NCC and SSIM (0.681) was found for registration. Data conclusion: Initial results demonstrate that the proposed method may be feasible in providing joint tumor segmentation and registration of MRI data acquired at different field strengths.
Evaluating Pre-trained Convolutional Neural Networks and Foundation Models as Feature Extractors for Content-based Medical Image Retrieval
Sep 14, 2024



Abstract:Medical image retrieval refers to the task of finding similar images for given query images in a database, with applications such as diagnosis support, treatment planning, and educational tools for inexperienced medical practitioners. While traditional medical image retrieval was performed using clinical metadata, content-based medical image retrieval (CBMIR) relies on the characteristic features of the images, such as color, texture, shape, and spatial features. Many approaches have been proposed for CBMIR, and among them, using pre-trained convolutional neural networks (CNNs) is a widely utilized approach. However, considering the recent advances in the development of foundation models for various computer vision tasks, their application for CBMIR can be also investigated for its potentially superior performance. In this study, we used several pre-trained feature extractors from well-known pre-trained CNNs (VGG19, ResNet-50, DenseNet121, and EfficientNetV2M) and pre-trained foundation models (MedCLIP, BioMedCLIP, OpenCLIP, CONCH and UNI) and investigated the CBMIR performance on a subset of the MedMNIST V2 dataset, including eight types of 2D and 3D medical images. Furthermore, we also investigated the effect of image size on the CBMIR performance. Our results show that, overall, for the 2D datasets, foundation models deliver superior performance by a large margin compared to CNNs, with UNI providing the best overall performance across all datasets and image sizes. For 3D datasets, CNNs and foundation models deliver more competitive performance, with CONCH achieving the best overall performance. Moreover, our findings confirm that while using larger image sizes (especially for 2D datasets) yields slightly better performance, competitive CBMIR performance can still be achieved even with smaller image sizes. Our codes to generate and reproduce the results are available on GitHub.
WCEbleedGen: A wireless capsule endoscopy dataset and its benchmarking for automatic bleeding classification, detection, and segmentation
Aug 22, 2024



Abstract:Computer-based analysis of Wireless Capsule Endoscopy (WCE) is crucial. However, a medically annotated WCE dataset for training and evaluation of automatic classification, detection, and segmentation of bleeding and non-bleeding frames is currently lacking. The present work focused on development of a medically annotated WCE dataset called WCEbleedGen for automatic classification, detection, and segmentation of bleeding and non-bleeding frames. It comprises 2,618 WCE bleeding and non-bleeding frames which were collected from various internet resources and existing WCE datasets. A comprehensive benchmarking and evaluation of the developed dataset was done using nine classification-based, three detection-based, and three segmentation-based deep learning models. The dataset is of high-quality, is class-balanced and contains single and multiple bleeding sites. Overall, our standard benchmark results show that Visual Geometric Group (VGG) 19, You Only Look Once version 8 nano (YOLOv8n), and Link network (Linknet) performed best in automatic classification, detection, and segmentation-based evaluations, respectively. Automatic bleeding diagnosis is crucial for WCE video interpretations. This diverse dataset will aid in developing of real-time, multi-task learning-based innovative solutions for automatic bleeding diagnosis in WCE. The dataset and code are publicly available at https://zenodo.org/records/10156571 and https://github.com/misahub2023/Benchmarking-Codes-of-the-WCEBleedGen-dataset.
Capsule Vision 2024 Challenge: Multi-Class Abnormality Classification for Video Capsule Endoscopy
Aug 09, 2024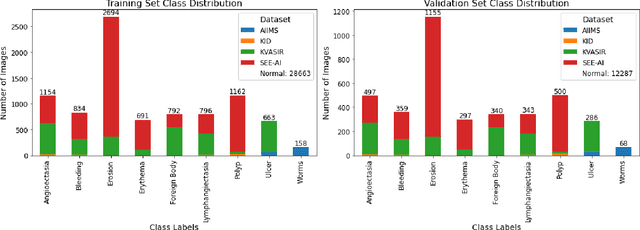
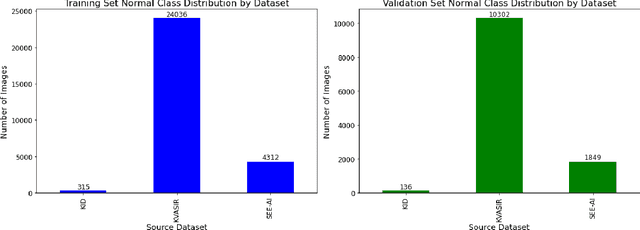
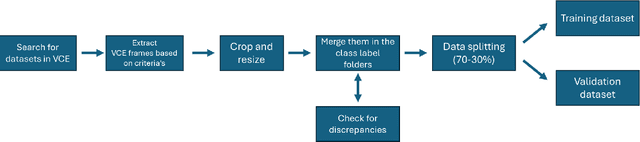
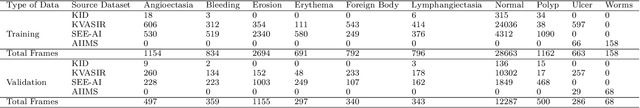
Abstract:We present the Capsule Vision 2024 Challenge: Multi-Class Abnormality Classification for Video Capsule Endoscopy. It is being virtually organized by the Research Center for Medical Image Analysis and Artificial Intelligence (MIAAI), Department of Medicine, Danube Private University, Krems, Austria and Medical Imaging and Signal Analysis Hub (MISAHUB) in collaboration with the 9th International Conference on Computer Vision & Image Processing (CVIP 2024) being organized by the Indian Institute of Information Technology, Design and Manufacturing (IIITDM) Kancheepuram, Chennai, India. This document describes the overview of the challenge, its registration and rules, submission format, and the description of the utilized datasets.
A Self-Supervised Image Registration Approach for Measuring Local Response Patterns in Metastatic Ovarian Cancer
Jul 24, 2024



Abstract:High-grade serous ovarian carcinoma (HGSOC) is characterised by significant spatial and temporal heterogeneity, typically manifesting at an advanced metastatic stage. A major challenge in treating advanced HGSOC is effectively monitoring localised change in tumour burden across multiple sites during neoadjuvant chemotherapy (NACT) and predicting long-term pathological response and overall patient survival. In this work, we propose a self-supervised deformable image registration algorithm that utilises a general-purpose image encoder for image feature extraction to co-register contrast-enhanced computerised tomography scan images acquired before and after neoadjuvant chemotherapy. This approach addresses challenges posed by highly complex tumour deformations and longitudinal lesion matching during treatment. Localised tumour changes are calculated using the Jacobian determinant maps of the registration deformation at multiple disease sites and their macroscopic areas, including hypo-dense (i.e., cystic/necrotic), hyper-dense (i.e., calcified), and intermediate density (i.e., soft tissue) portions. A series of experiments is conducted to understand the role of a general-purpose image encoder and its application in quantifying change in tumour burden during neoadjuvant chemotherapy in HGSOC. This work is the first to demonstrate the feasibility of a self-supervised image registration approach in quantifying NACT-induced localised tumour changes across the whole disease burden of patients with complex multi-site HGSOC, which could be used as a potential marker for ovarian cancer patient's long-term pathological response and survival.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge