Leonardo Rundo
A Self-Supervised Image Registration Approach for Measuring Local Response Patterns in Metastatic Ovarian Cancer
Jul 24, 2024



Abstract:High-grade serous ovarian carcinoma (HGSOC) is characterised by significant spatial and temporal heterogeneity, typically manifesting at an advanced metastatic stage. A major challenge in treating advanced HGSOC is effectively monitoring localised change in tumour burden across multiple sites during neoadjuvant chemotherapy (NACT) and predicting long-term pathological response and overall patient survival. In this work, we propose a self-supervised deformable image registration algorithm that utilises a general-purpose image encoder for image feature extraction to co-register contrast-enhanced computerised tomography scan images acquired before and after neoadjuvant chemotherapy. This approach addresses challenges posed by highly complex tumour deformations and longitudinal lesion matching during treatment. Localised tumour changes are calculated using the Jacobian determinant maps of the registration deformation at multiple disease sites and their macroscopic areas, including hypo-dense (i.e., cystic/necrotic), hyper-dense (i.e., calcified), and intermediate density (i.e., soft tissue) portions. A series of experiments is conducted to understand the role of a general-purpose image encoder and its application in quantifying change in tumour burden during neoadjuvant chemotherapy in HGSOC. This work is the first to demonstrate the feasibility of a self-supervised image registration approach in quantifying NACT-induced localised tumour changes across the whole disease burden of patients with complex multi-site HGSOC, which could be used as a potential marker for ovarian cancer patient's long-term pathological response and survival.
Calibrating Ensembles for Scalable Uncertainty Quantification in Deep Learning-based Medical Segmentation
Sep 20, 2022
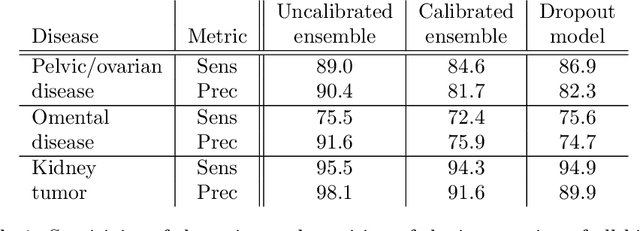


Abstract:Uncertainty quantification in automated image analysis is highly desired in many applications. Typically, machine learning models in classification or segmentation are only developed to provide binary answers; however, quantifying the uncertainty of the models can play a critical role for example in active learning or machine human interaction. Uncertainty quantification is especially difficult when using deep learning-based models, which are the state-of-the-art in many imaging applications. The current uncertainty quantification approaches do not scale well in high-dimensional real-world problems. Scalable solutions often rely on classical techniques, such as dropout, during inference or training ensembles of identical models with different random seeds to obtain a posterior distribution. In this paper, we show that these approaches fail to approximate the classification probability. On the contrary, we propose a scalable and intuitive framework to calibrate ensembles of deep learning models to produce uncertainty quantification measurements that approximate the classification probability. On unseen test data, we demonstrate improved calibration, sensitivity (in two out of three cases) and precision when being compared with the standard approaches. We further motivate the usage of our method in active learning, creating pseudo-labels to learn from unlabeled images and human-machine collaboration.
Focal Attention Networks: optimising attention for biomedical image segmentation
Oct 31, 2021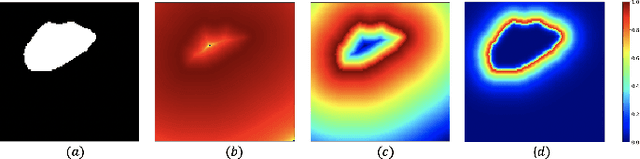
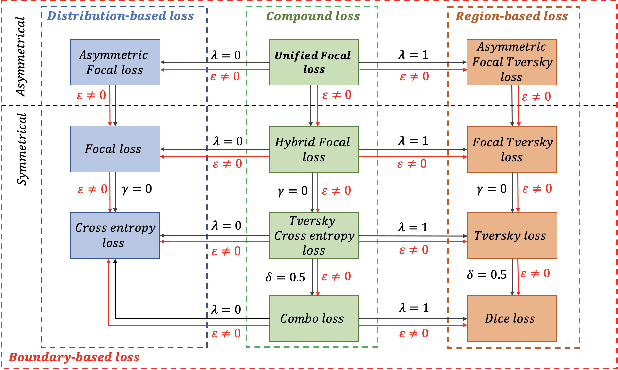
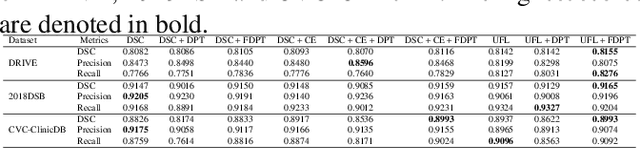
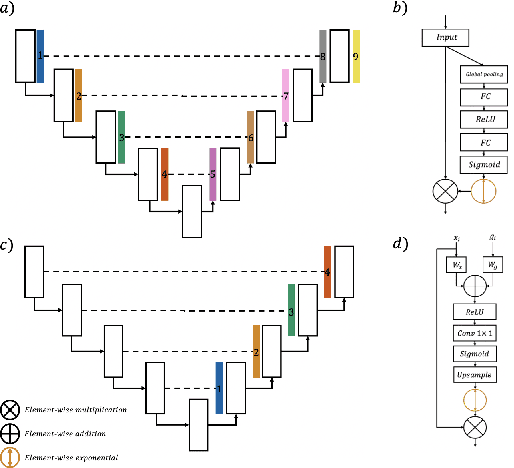
Abstract:In recent years, there has been increasing interest to incorporate attention into deep learning architectures for biomedical image segmentation. The modular design of attention mechanisms enables flexible integration into convolutional neural network architectures, such as the U-Net. Whether attention is appropriate to use, what type of attention to use, and where in the network to incorporate attention modules, are all important considerations that are currently overlooked. In this paper, we investigate the role of the Focal parameter in modulating attention, revealing a link between attention in loss functions and networks. By incorporating a Focal distance penalty term, we extend the Unified Focal loss framework to include boundary-based losses. Furthermore, we develop a simple and interpretable, dataset and model-specific heuristic to integrate the Focal parameter into the Squeeze-and-Excitation block and Attention Gate, achieving optimal performance with fewer number of attention modules on three well-validated biomedical imaging datasets, suggesting judicious use of attention modules results in better performance and efficiency.
Incorporating Boundary Uncertainty into loss functions for biomedical image segmentation
Oct 31, 2021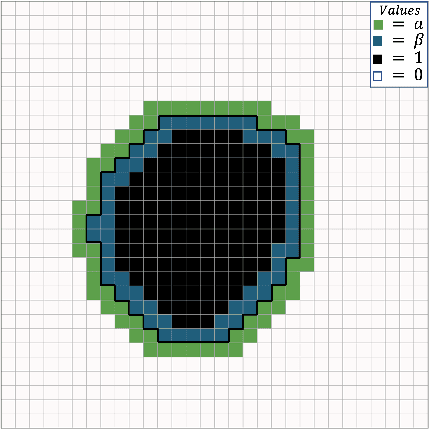


Abstract:Manual segmentation is used as the gold-standard for evaluating neural networks on automated image segmentation tasks. Due to considerable heterogeneity in shapes, colours and textures, demarcating object boundaries is particularly difficult in biomedical images, resulting in significant inter and intra-rater variability. Approaches, such as soft labelling and distance penalty term, apply a global transformation to the ground truth, redefining the loss function with respect to uncertainty. However, global operations are computationally expensive, and neither approach accurately reflects the uncertainty underlying manual annotation. In this paper, we propose the Boundary Uncertainty, which uses morphological operations to restrict soft labelling to object boundaries, providing an appropriate representation of uncertainty in ground truth labels, and may be adapted to enable robust model training where systematic manual segmentation errors are present. We incorporate Boundary Uncertainty with the Dice loss, achieving consistently improved performance across three well-validated biomedical imaging datasets compared to soft labelling and distance-weighted penalty. Boundary Uncertainty not only more accurately reflects the segmentation process, but it is also efficient, robust to segmentation errors and exhibits better generalisation.
Calibrating the Dice loss to handle neural network overconfidence for biomedical image segmentation
Oct 31, 2021



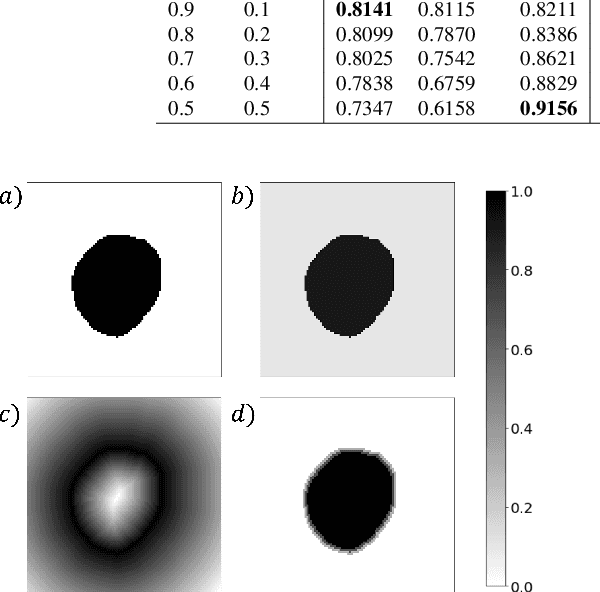
Abstract:The Dice similarity coefficient (DSC) is both a widely used metric and loss function for biomedical image segmentation due to its robustness to class imbalance. However, it is well known that the DSC loss is poorly calibrated, resulting in overconfident predictions that cannot be usefully interpreted in biomedical and clinical practice. Performance is often the only metric used to evaluate segmentations produced by deep neural networks, and calibration is often neglected. However, calibration is important for translation into biomedical and clinical practice, providing crucial contextual information to model predictions for interpretation by scientists and clinicians. In this study, we identify poor calibration as an emerging challenge of deep learning based biomedical image segmentation. We provide a simple yet effective extension of the DSC loss, named the DSC++ loss, that selectively modulates the penalty associated with overconfident, incorrect predictions. As a standalone loss function, the DSC++ loss achieves significantly improved calibration over the conventional DSC loss across five well-validated open-source biomedical imaging datasets. Similarly, we observe significantly improved when integrating the DSC++ loss into four DSC-based loss functions. Finally, we use softmax thresholding to illustrate that well calibrated outputs enable tailoring of precision-recall bias, an important post-processing technique to adapt the model predictions to suit the biomedical or clinical task. The DSC++ loss overcomes the major limitation of the DSC, providing a suitable loss function for training deep learning segmentation models for use in biomedical and clinical practice.
Computer-Assisted Analysis of Biomedical Images
Jun 04, 2021Abstract:Nowadays, the amount of heterogeneous biomedical data is increasing more and more thanks to novel sensing techniques and high-throughput technologies. In reference to biomedical image analysis, the advances in image acquisition modalities and high-throughput imaging experiments are creating new challenges. This huge information ensemble could overwhelm the analytic capabilities needed by physicians in their daily decision-making tasks as well as by biologists investigating complex biochemical systems. In particular, quantitative imaging methods convey scientifically and clinically relevant information in prediction, prognosis or treatment response assessment, by also considering radiomics approaches. Therefore, the computational analysis of medical and biological images plays a key role in radiology and laboratory applications. In this regard, frameworks based on advanced Machine Learning and Computational Intelligence can significantly improve traditional Image Processing and Pattern Recognition approaches. However, conventional Artificial Intelligence techniques must be tailored to address the unique challenges concerning biomedical imaging data. This thesis aims at proposing novel and advanced computer-assisted methods for biomedical image analysis, also as an instrument in the development of Clinical Decision Support Systems, by always keeping in mind the clinical feasibility of the developed solutions. In conclusion, the ultimate goal of these research studies is to gain clinically and biologically useful insights that can guide differential diagnosis and therapies, leading towards biomedical data integration for personalized medicine. As a matter of fact, the proposed computer-assisted bioimage analysis methods can be beneficial for the definition of imaging biomarkers, as well as for quantitative medicine and biology.
Advances in Artificial Intelligence to Reduce Polyp Miss Rates during Colonoscopy
May 16, 2021
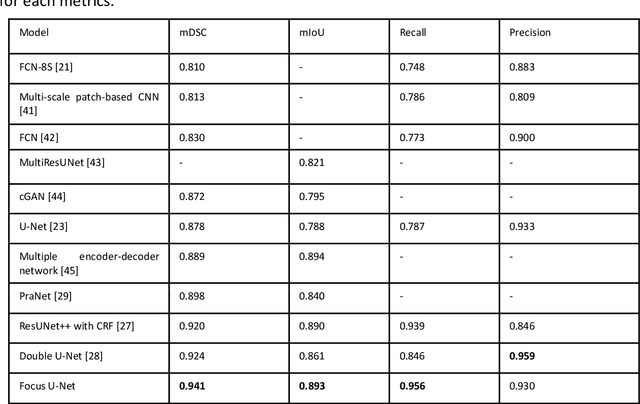
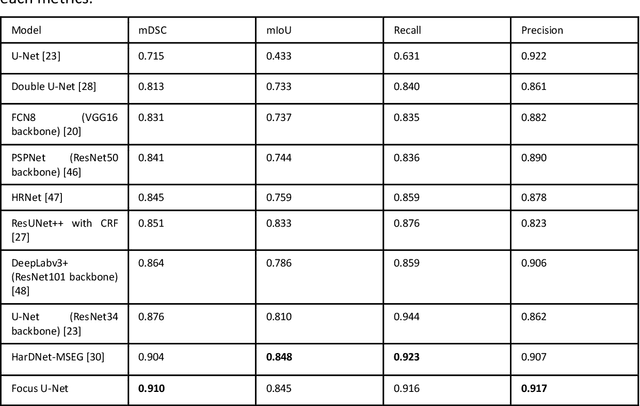

Abstract:BACKGROUND AND CONTEXT: Artificial intelligence has the potential to aid gastroenterologists by reducing polyp miss detection rates during colonoscopy screening for colorectal cancer. NEW FINDINGS: We introduce a new deep neural network architecture, the Focus U-Net, which achieves state-of-the-art performance for polyp segmentation across five public datasets containing images of polyps obtained during colonoscopy. LIMITATIONS: The model has been validated on images taken during colonoscopy but requires validation on live video data to ensure generalisability. IMPACT: Once validated on live video data, our polyp segmentation algorithm could be integrated into colonoscopy practice and assist gastroenterologists by reducing the number of polyps missed
A Mixed Focal Loss Function for Handling Class Imbalanced Medical Image Segmentation
Feb 08, 2021



Abstract:Automatic segmentation methods are an important advancement in medical imaging analysis. Machine learning techniques, and deep neural networks in particular, are the state-of-the-art for most automated medical image segmentation tasks, ranging from the subcellular to the level of organ systems. Issues with class imbalance pose a significant challenge irrespective of scale, with organs, and especially with tumours, often occupying a considerably smaller volume relative to the background. Loss functions used in the training of segmentation algorithms differ in their robustness to class imbalance, with cross entropy-based losses being more affected than Dice-based losses. In this work, we first experiment with seven different Dice-based and cross entropy-based loss functions on the publicly available Kidney Tumour Segmentation 2019 (KiTS19) Computed Tomography dataset, and then further evaluate the top three performing loss functions on the Brain Tumour Segmentation 2020 (BraTS20) Magnetic Resonance Imaging dataset. Motivated by the results of our study, we propose a Mixed Focal loss function, a new compound loss function derived from modified variants of the Focal loss and Focal Dice loss functions. We demonstrate that our proposed loss function is associated with a better recall-precision balance, significantly outperforming the other loss functions in both binary and multi-class image segmentation. Importantly, the proposed Mixed Focal loss function is robust to significant class imbalance. Furthermore, we showed the benefit of using compound losses over their component losses, and the improvement provided by the focal variants over other variants.
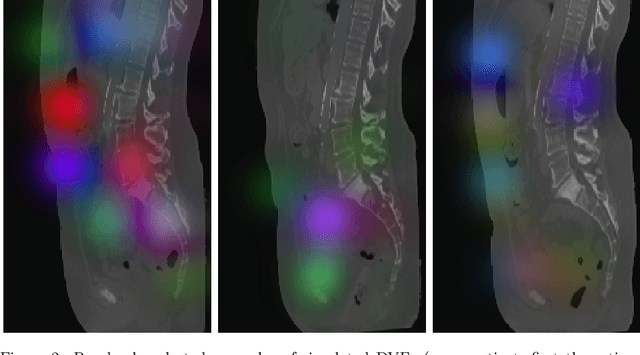
MADGAN: unsupervised Medical Anomaly Detection GAN using multiple adjacent brain MRI slice reconstruction
Jul 24, 2020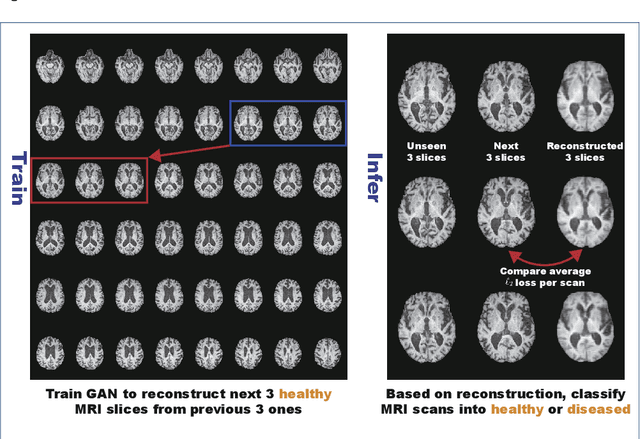
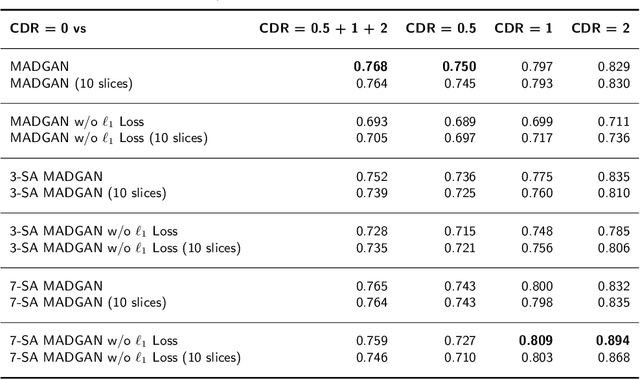
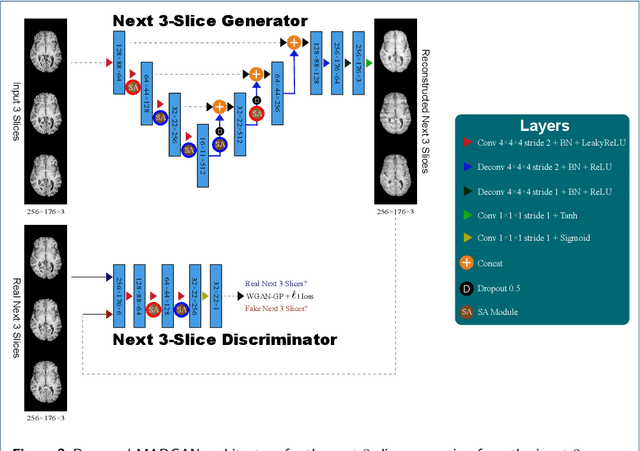
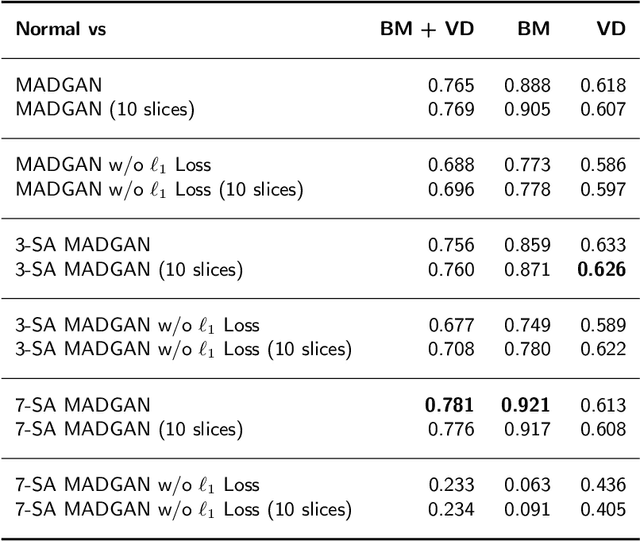
Abstract:Unsupervised learning can discover various unseen diseases, relying on large-scale unannotated medical images of healthy subjects. Towards this, unsupervised methods reconstruct a 2D/3D single medical image to detect outliers either in the learned feature space or from high reconstruction loss. However, without considering continuity between multiple adjacent slices, they cannot directly discriminate diseases composed of the accumulation of subtle anatomical anomalies, such as Alzheimer's Disease (AD). Moreover, no study has shown how unsupervised anomaly detection is associated with either disease stages, various (i.e., more than two types of) diseases, or multi-sequence Magnetic Resonance Imaging (MRI) scans. Therefore, we propose unsupervised Medical Anomaly Detection Generative Adversarial Network (MADGAN), a novel two-step method using GAN-based multiple adjacent brain MRI slice reconstruction to detect various diseases at different stages on multi-sequence structural MRI: (Reconstruction) Wasserstein loss with Gradient Penalty + 100 L1 loss-trained on 3 healthy brain axial MRI slices to reconstruct the next 3 ones-reconstructs unseen healthy/abnormal scans; (Diagnosis) Average L2 loss per scan discriminates them, comparing the ground truth/reconstructed slices. For training, we use 1,133 healthy T1-weighted (T1) and 135 healthy contrast-enhanced T1 (T1c) brain MRI scans. Our Self-Attention MADGAN can detect AD on T1 scans at a very early stage, Mild Cognitive Impairment (MCI), with Area Under the Curve (AUC) 0.727, and AD at a late stage with AUC 0.894, while detecting brain metastases on T1c scans with AUC 0.921.
3D deformable registration of longitudinal abdominopelvic CT images using unsupervised deep learning
May 15, 2020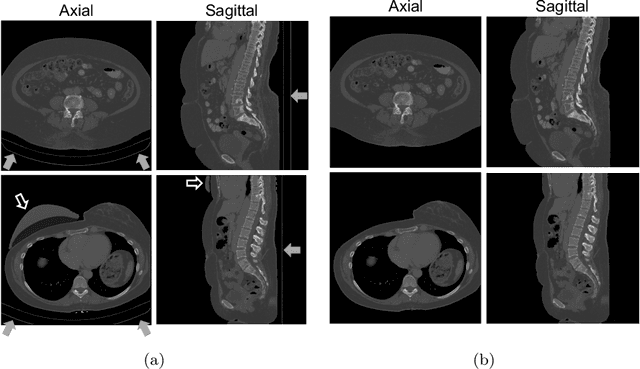

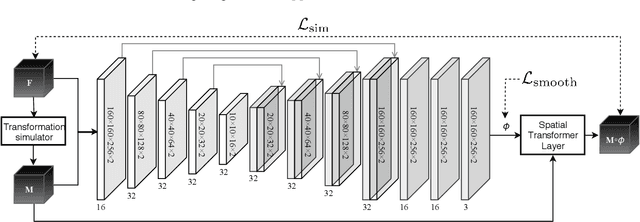
Abstract:This study investigates the use of the unsupervised deep learning framework VoxelMorph for deformable registration of longitudinal abdominopelvic CT images acquired in patients with bone metastases from breast cancer. The CT images were refined prior to registration by automatically removing the CT table and all other extra-corporeal components. To improve the learning capabilities of VoxelMorph when only a limited amount of training data is available, a novel incremental training strategy is proposed based on simulated deformations of consecutive CT images. In a 4-fold cross-validation scheme, the incremental training strategy achieved significantly better registration performance compared to training on a single volume. Although our deformable image registration method did not outperform iterative registration using NiftyReg (considered as a benchmark) in terms of registration quality, the registrations were approximately 300 times faster. This study showed the feasibility of deep learning based deformable registration of longitudinal abdominopelvic CT images via a novel incremental training strategy based on simulated deformations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge