Tomoyuki Noguchi
MADGAN: unsupervised Medical Anomaly Detection GAN using multiple adjacent brain MRI slice reconstruction
Jul 24, 2020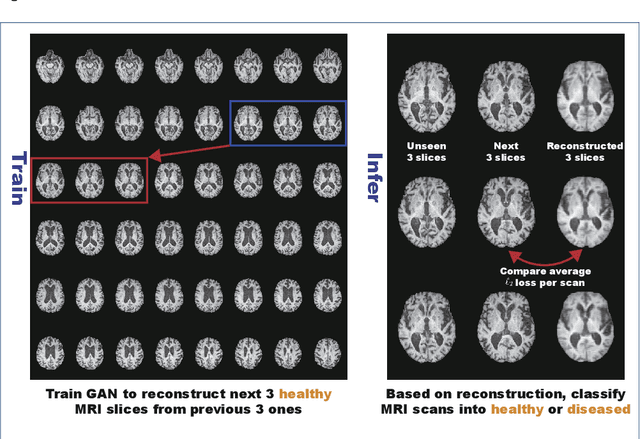
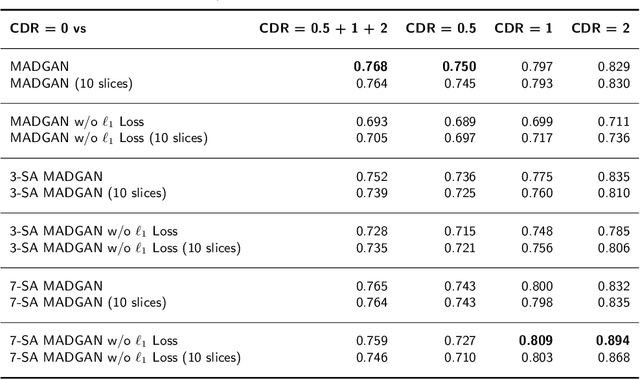
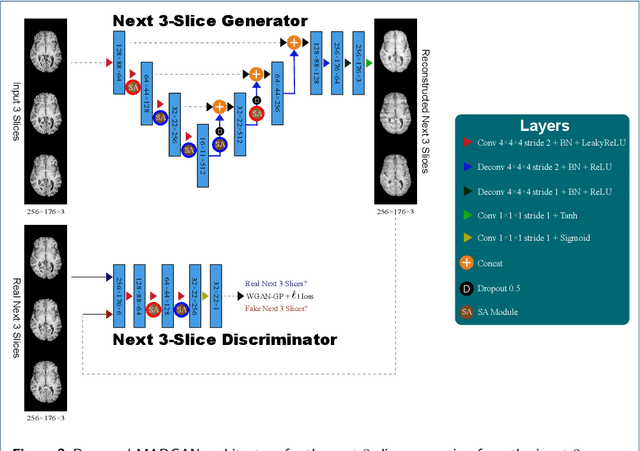
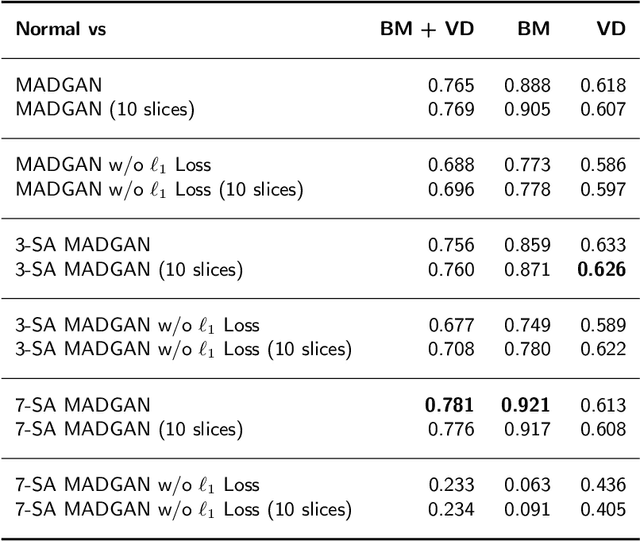
Abstract:Unsupervised learning can discover various unseen diseases, relying on large-scale unannotated medical images of healthy subjects. Towards this, unsupervised methods reconstruct a 2D/3D single medical image to detect outliers either in the learned feature space or from high reconstruction loss. However, without considering continuity between multiple adjacent slices, they cannot directly discriminate diseases composed of the accumulation of subtle anatomical anomalies, such as Alzheimer's Disease (AD). Moreover, no study has shown how unsupervised anomaly detection is associated with either disease stages, various (i.e., more than two types of) diseases, or multi-sequence Magnetic Resonance Imaging (MRI) scans. Therefore, we propose unsupervised Medical Anomaly Detection Generative Adversarial Network (MADGAN), a novel two-step method using GAN-based multiple adjacent brain MRI slice reconstruction to detect various diseases at different stages on multi-sequence structural MRI: (Reconstruction) Wasserstein loss with Gradient Penalty + 100 L1 loss-trained on 3 healthy brain axial MRI slices to reconstruct the next 3 ones-reconstructs unseen healthy/abnormal scans; (Diagnosis) Average L2 loss per scan discriminates them, comparing the ground truth/reconstructed slices. For training, we use 1,133 healthy T1-weighted (T1) and 135 healthy contrast-enhanced T1 (T1c) brain MRI scans. Our Self-Attention MADGAN can detect AD on T1 scans at a very early stage, Mild Cognitive Impairment (MCI), with Area Under the Curve (AUC) 0.727, and AD at a late stage with AUC 0.894, while detecting brain metastases on T1c scans with AUC 0.921.
Learning More with Less: Conditional PGGAN-based Data Augmentation for Brain Metastases Detection Using Highly-Rough Annotation on MR Images
Mar 03, 2019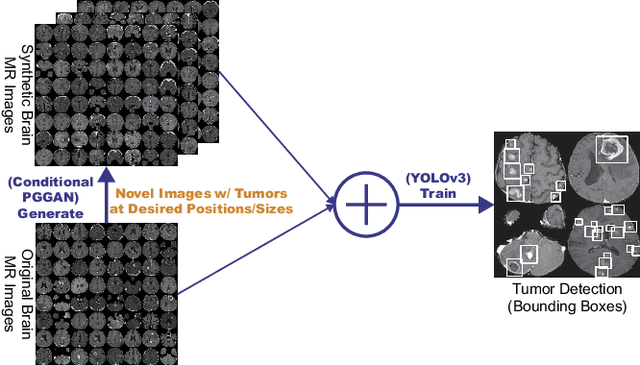
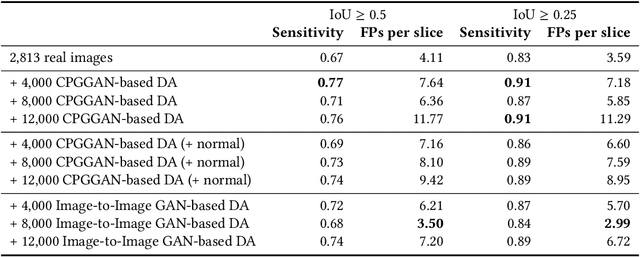
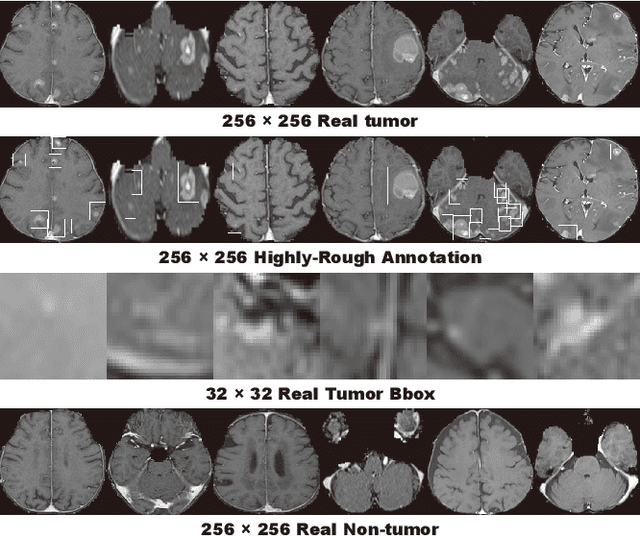
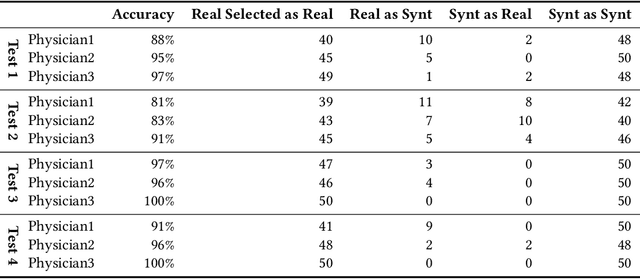
Abstract:Accurate computer-assisted diagnosis can alleviate the risk of overlooking the diagnosis in a clinical environment. Towards this, as a Data Augmentation (DA) technique, Generative Adversarial Networks (GANs) can synthesize additional training data to handle small/fragmented medical images from various scanners; those images are realistic but completely different from the original ones, filling the data lack in the real image distribution. However, we cannot easily use them to locate the position of disease areas, considering expert physicians' annotation as time-expensive tasks. Therefore, this paper proposes Conditional Progressive Growing of GANs (CPGGANs), incorporating bounding box conditions into PGGANs to place brain metastases at desired position/size on 256 x 256 Magnetic Resonance (MR) images, for Convolutional Neural Network-based tumor detection; this first GAN-based medical DA using automatic bounding box annotation improves the robustness during training. The results show that CPGGAN-based DA can boost 10% sensitivity in diagnosis with an acceptable amount of additional False Positives---even with physicians' highly-rough and inconsistent bounding box annotation. Surprisingly, further realistic tumor appearance, achieved with additional normal brain MR images for CPGGAN training, does not contribute to detection performance, while even three expert physicians cannot accurately distinguish them from the real ones in Visual Turing Test.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge