Michael Yeung
on behalf of the AIX-COVNET collaboration
VariFace: Fair and Diverse Synthetic Dataset Generation for Face Recognition
Dec 09, 2024



Abstract:The use of large-scale, web-scraped datasets to train face recognition models has raised significant privacy and bias concerns. Synthetic methods mitigate these concerns and provide scalable and controllable face generation to enable fair and accurate face recognition. However, existing synthetic datasets display limited intraclass and interclass diversity and do not match the face recognition performance obtained using real datasets. Here, we propose VariFace, a two-stage diffusion-based pipeline to create fair and diverse synthetic face datasets to train face recognition models. Specifically, we introduce three methods: Face Recognition Consistency to refine demographic labels, Face Vendi Score Guidance to improve interclass diversity, and Divergence Score Conditioning to balance the identity preservation-intraclass diversity trade-off. When constrained to the same dataset size, VariFace considerably outperforms previous synthetic datasets (0.9200 $\rightarrow$ 0.9405) and achieves comparable performance to face recognition models trained with real data (Real Gap = -0.0065). In an unconstrained setting, VariFace not only consistently achieves better performance compared to previous synthetic methods across dataset sizes but also, for the first time, outperforms the real dataset (CASIA-WebFace) across six evaluation datasets. This sets a new state-of-the-art performance with an average face verification accuracy of 0.9567 (Real Gap = +0.0097) across LFW, CFP-FP, CPLFW, AgeDB, and CALFW datasets and 0.9366 (Real Gap = +0.0380) on the RFW dataset.
Stain Consistency Learning: Handling Stain Variation for Automatic Digital Pathology Segmentation
Nov 11, 2023Abstract:Stain variation is a unique challenge associated with automated analysis of digital pathology. Numerous methods have been developed to improve the robustness of machine learning methods to stain variation, but comparative studies have demonstrated limited benefits to performance. Moreover, methods to handle stain variation were largely developed for H&E stained data, with evaluation generally limited to classification tasks. Here we propose Stain Consistency Learning, a novel framework combining stain-specific augmentation with a stain consistency loss function to learn stain colour invariant features. We perform the first, extensive comparison of methods to handle stain variation for segmentation tasks, comparing ten methods on Masson's trichrome and H&E stained cell and nuclei datasets, respectively. We observed that stain normalisation methods resulted in equivalent or worse performance, while stain augmentation or stain adversarial methods demonstrated improved performance, with the best performance consistently achieved by our proposed approach. The code is available at: https://github.com/mlyg/stain_consistency_learning
Focal Attention Networks: optimising attention for biomedical image segmentation
Oct 31, 2021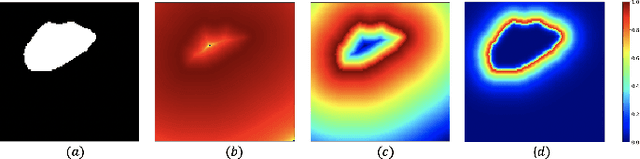
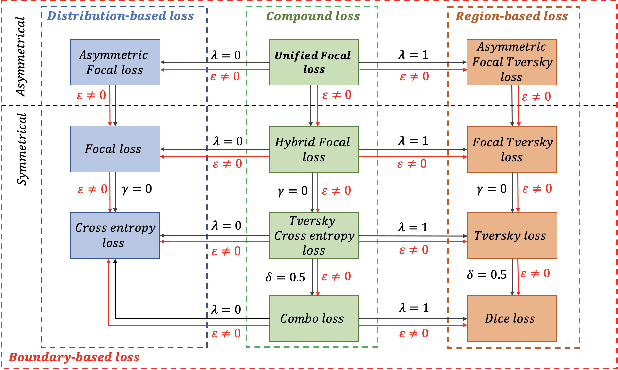
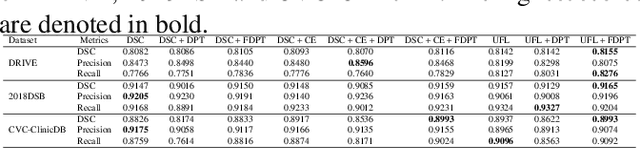
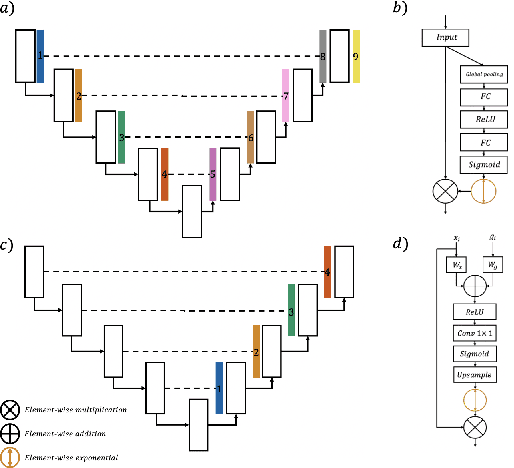
Abstract:In recent years, there has been increasing interest to incorporate attention into deep learning architectures for biomedical image segmentation. The modular design of attention mechanisms enables flexible integration into convolutional neural network architectures, such as the U-Net. Whether attention is appropriate to use, what type of attention to use, and where in the network to incorporate attention modules, are all important considerations that are currently overlooked. In this paper, we investigate the role of the Focal parameter in modulating attention, revealing a link between attention in loss functions and networks. By incorporating a Focal distance penalty term, we extend the Unified Focal loss framework to include boundary-based losses. Furthermore, we develop a simple and interpretable, dataset and model-specific heuristic to integrate the Focal parameter into the Squeeze-and-Excitation block and Attention Gate, achieving optimal performance with fewer number of attention modules on three well-validated biomedical imaging datasets, suggesting judicious use of attention modules results in better performance and efficiency.
Incorporating Boundary Uncertainty into loss functions for biomedical image segmentation
Oct 31, 2021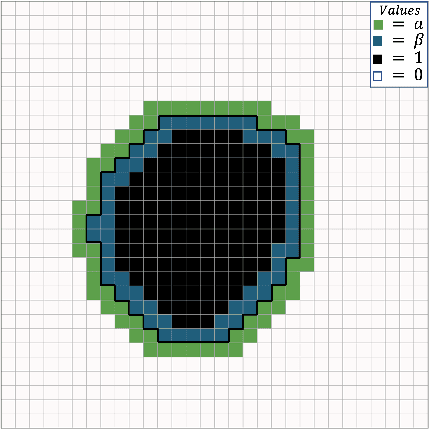

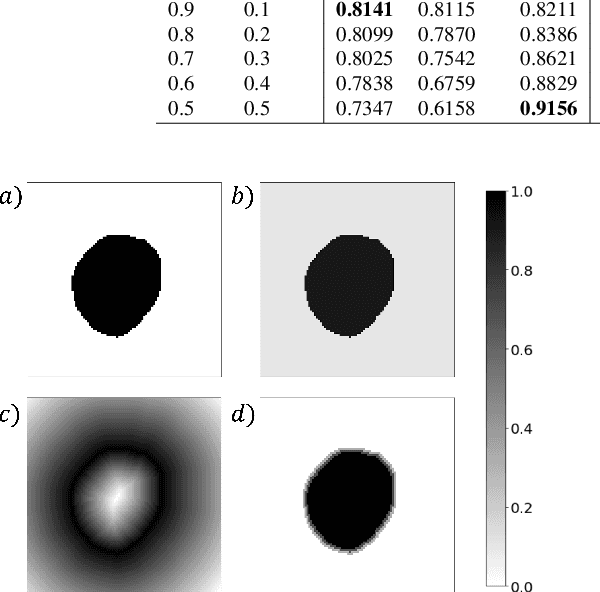

Abstract:Manual segmentation is used as the gold-standard for evaluating neural networks on automated image segmentation tasks. Due to considerable heterogeneity in shapes, colours and textures, demarcating object boundaries is particularly difficult in biomedical images, resulting in significant inter and intra-rater variability. Approaches, such as soft labelling and distance penalty term, apply a global transformation to the ground truth, redefining the loss function with respect to uncertainty. However, global operations are computationally expensive, and neither approach accurately reflects the uncertainty underlying manual annotation. In this paper, we propose the Boundary Uncertainty, which uses morphological operations to restrict soft labelling to object boundaries, providing an appropriate representation of uncertainty in ground truth labels, and may be adapted to enable robust model training where systematic manual segmentation errors are present. We incorporate Boundary Uncertainty with the Dice loss, achieving consistently improved performance across three well-validated biomedical imaging datasets compared to soft labelling and distance-weighted penalty. Boundary Uncertainty not only more accurately reflects the segmentation process, but it is also efficient, robust to segmentation errors and exhibits better generalisation.
Calibrating the Dice loss to handle neural network overconfidence for biomedical image segmentation
Oct 31, 2021



Abstract:The Dice similarity coefficient (DSC) is both a widely used metric and loss function for biomedical image segmentation due to its robustness to class imbalance. However, it is well known that the DSC loss is poorly calibrated, resulting in overconfident predictions that cannot be usefully interpreted in biomedical and clinical practice. Performance is often the only metric used to evaluate segmentations produced by deep neural networks, and calibration is often neglected. However, calibration is important for translation into biomedical and clinical practice, providing crucial contextual information to model predictions for interpretation by scientists and clinicians. In this study, we identify poor calibration as an emerging challenge of deep learning based biomedical image segmentation. We provide a simple yet effective extension of the DSC loss, named the DSC++ loss, that selectively modulates the penalty associated with overconfident, incorrect predictions. As a standalone loss function, the DSC++ loss achieves significantly improved calibration over the conventional DSC loss across five well-validated open-source biomedical imaging datasets. Similarly, we observe significantly improved when integrating the DSC++ loss into four DSC-based loss functions. Finally, we use softmax thresholding to illustrate that well calibrated outputs enable tailoring of precision-recall bias, an important post-processing technique to adapt the model predictions to suit the biomedical or clinical task. The DSC++ loss overcomes the major limitation of the DSC, providing a suitable loss function for training deep learning segmentation models for use in biomedical and clinical practice.
Advances in Artificial Intelligence to Reduce Polyp Miss Rates during Colonoscopy
May 16, 2021
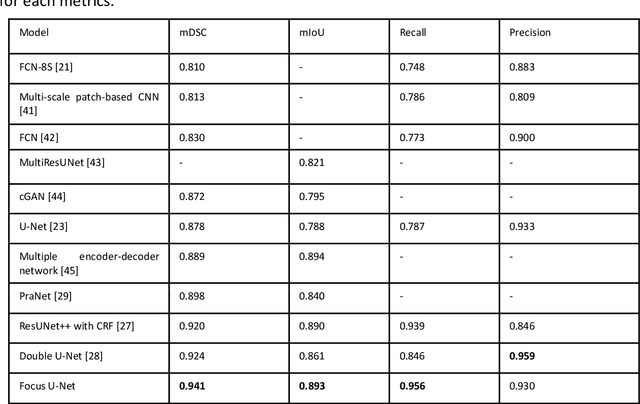
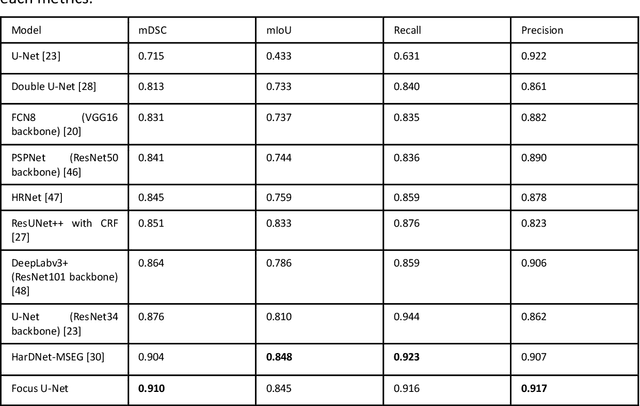

Abstract:BACKGROUND AND CONTEXT: Artificial intelligence has the potential to aid gastroenterologists by reducing polyp miss detection rates during colonoscopy screening for colorectal cancer. NEW FINDINGS: We introduce a new deep neural network architecture, the Focus U-Net, which achieves state-of-the-art performance for polyp segmentation across five public datasets containing images of polyps obtained during colonoscopy. LIMITATIONS: The model has been validated on images taken during colonoscopy but requires validation on live video data to ensure generalisability. IMPACT: Once validated on live video data, our polyp segmentation algorithm could be integrated into colonoscopy practice and assist gastroenterologists by reducing the number of polyps missed
A Mixed Focal Loss Function for Handling Class Imbalanced Medical Image Segmentation
Feb 08, 2021



Abstract:Automatic segmentation methods are an important advancement in medical imaging analysis. Machine learning techniques, and deep neural networks in particular, are the state-of-the-art for most automated medical image segmentation tasks, ranging from the subcellular to the level of organ systems. Issues with class imbalance pose a significant challenge irrespective of scale, with organs, and especially with tumours, often occupying a considerably smaller volume relative to the background. Loss functions used in the training of segmentation algorithms differ in their robustness to class imbalance, with cross entropy-based losses being more affected than Dice-based losses. In this work, we first experiment with seven different Dice-based and cross entropy-based loss functions on the publicly available Kidney Tumour Segmentation 2019 (KiTS19) Computed Tomography dataset, and then further evaluate the top three performing loss functions on the Brain Tumour Segmentation 2020 (BraTS20) Magnetic Resonance Imaging dataset. Motivated by the results of our study, we propose a Mixed Focal loss function, a new compound loss function derived from modified variants of the Focal loss and Focal Dice loss functions. We demonstrate that our proposed loss function is associated with a better recall-precision balance, significantly outperforming the other loss functions in both binary and multi-class image segmentation. Importantly, the proposed Mixed Focal loss function is robust to significant class imbalance. Furthermore, we showed the benefit of using compound losses over their component losses, and the improvement provided by the focal variants over other variants.
Machine learning for COVID-19 detection and prognostication using chest radiographs and CT scans: a systematic methodological review
Sep 01, 2020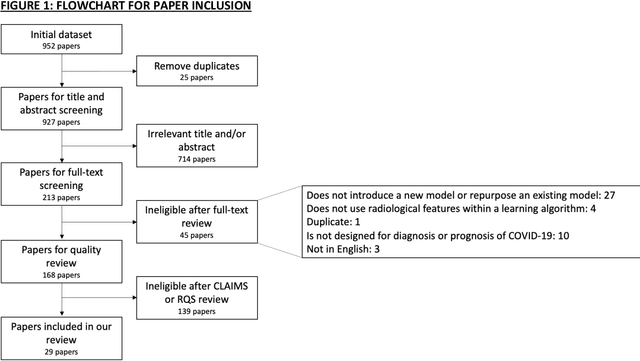
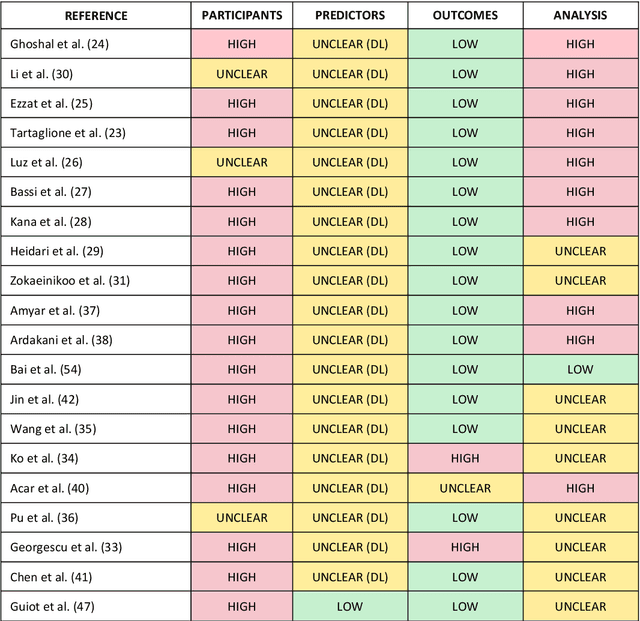
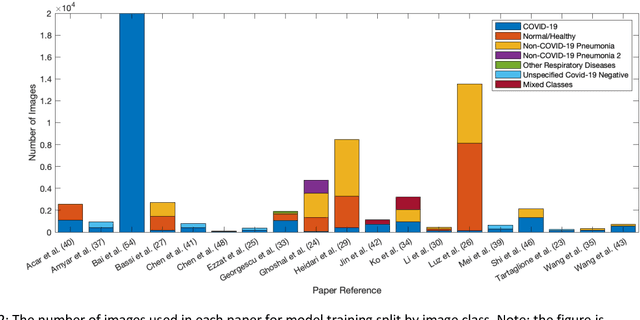
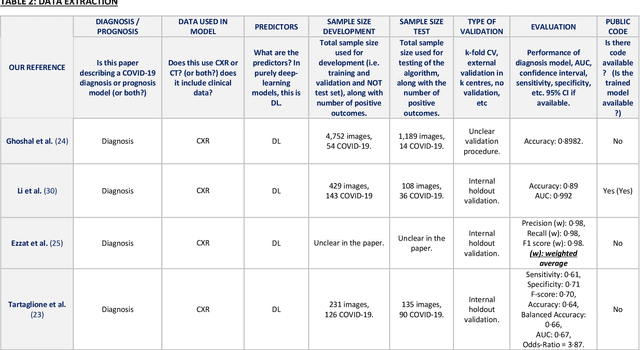
Abstract:Background: Machine learning methods offer great potential for fast and accurate detection and prognostication of COVID-19 from standard-of-care chest radiographs (CXR) and computed tomography (CT) images. In this systematic review we critically evaluate the machine learning methodologies employed in the rapidly growing literature. Methods: In this systematic review we reviewed EMBASE via OVID, MEDLINE via PubMed, bioRxiv, medRxiv and arXiv for published papers and preprints uploaded from Jan 1, 2020 to June 24, 2020. Studies which consider machine learning models for the diagnosis or prognosis of COVID-19 from CXR or CT images were included. A methodology quality review of each paper was performed against established benchmarks to ensure the review focusses only on high-quality reproducible papers. This study is registered with PROSPERO [CRD42020188887]. Interpretation: Our review finds that none of the developed models discussed are of potential clinical use due to methodological flaws and underlying biases. This is a major weakness, given the urgency with which validated COVID-19 models are needed. Typically, we find that the documentation of a model's development is not sufficient to make the results reproducible and therefore of 168 candidate papers only 29 are deemed to be reproducible and subsequently considered in this review. We therefore encourage authors to use established machine learning checklists to ensure sufficient documentation is made available, and to follow the PROBAST (prediction model risk of bias assessment tool) framework to determine the underlying biases in their model development process and to mitigate these where possible. This is key to safe clinical implementation which is urgently needed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge