Felix Lucka
Towards robust quantitative photoacoustic tomography via learned iterative methods
Oct 31, 2025Abstract:Photoacoustic tomography (PAT) is a medical imaging modality that can provide high-resolution tissue images based on the optical absorption. Classical reconstruction methods for quantifying the absorption coefficients rely on sufficient prior information to overcome noisy and imperfect measurements. As these methods utilize computationally expensive forward models, the computation becomes slow, limiting their potential for time-critical applications. As an alternative approach, deep learning-based reconstruction methods have been established for faster and more accurate reconstructions. However, most of these methods rely on having a large amount of training data, which is not the case in practice. In this work, we adopt the model-based learned iterative approach for the use in Quantitative PAT (QPAT), in which additional information from the model is iteratively provided to the updating networks, allowing better generalizability with scarce training data. We compare the performance of different learned updates based on gradient descent, Gauss-Newton, and Quasi-Newton methods. The learning tasks are formulated as greedy, requiring iterate-wise optimality, as well as end-to-end, where all networks are trained jointly. The implemented methods are tested with ideal simulated data as well as against a digital twin dataset that emulates scarce training data and high modeling error.
Benchmarking learned algorithms for computed tomography image reconstruction tasks
Dec 11, 2024



Abstract:Computed tomography (CT) is a widely used non-invasive diagnostic method in various fields, and recent advances in deep learning have led to significant progress in CT image reconstruction. However, the lack of large-scale, open-access datasets has hindered the comparison of different types of learned methods. To address this gap, we use the 2DeteCT dataset, a real-world experimental computed tomography dataset, for benchmarking machine learning based CT image reconstruction algorithms. We categorize these methods into post-processing networks, learned/unrolled iterative methods, learned regularizer methods, and plug-and-play methods, and provide a pipeline for easy implementation and evaluation. Using key performance metrics, including SSIM and PSNR, our benchmarking results showcase the effectiveness of various algorithms on tasks such as full data reconstruction, limited-angle reconstruction, sparse-angle reconstruction, low-dose reconstruction, and beam-hardening corrected reconstruction. With this benchmarking study, we provide an evaluation of a range of algorithms representative for different categories of learned reconstruction methods on a recently published dataset of real-world experimental CT measurements. The reproducible setup of methods and CT image reconstruction tasks in an open-source toolbox enables straightforward addition and comparison of new methods later on. The toolbox also provides the option to load the 2DeteCT dataset differently for extensions to other problems and different CT reconstruction tasks.
Exploring Out-of-distribution Detection for Sparse-view Computed Tomography with Diffusion Models
Nov 09, 2024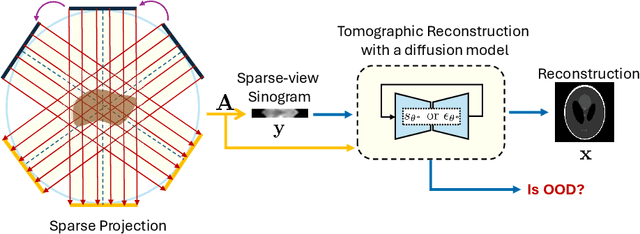
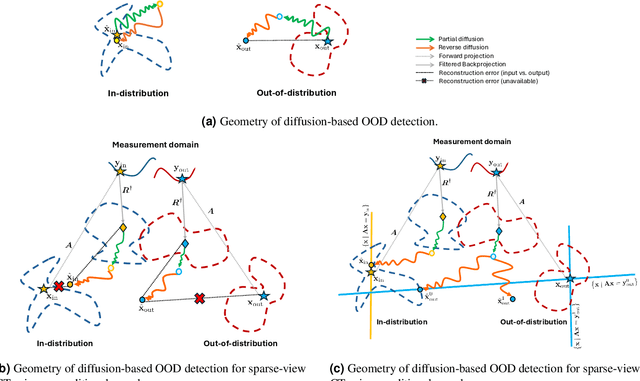
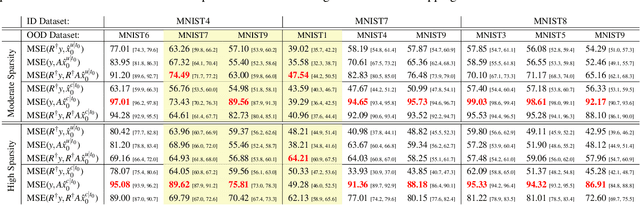

Abstract:Recent works demonstrate the effectiveness of diffusion models as unsupervised solvers for inverse imaging problems. Sparse-view computed tomography (CT) has greatly benefited from these advancements, achieving improved generalization without reliance on measurement parameters. However, this comes at the cost of potential hallucinations, especially when handling out-of-distribution (OOD) data. To ensure reliability, it is essential to study OOD detection for CT reconstruction across both clinical and industrial applications. This need further extends to enabling the OOD detector to function effectively as an anomaly inspection tool. In this paper, we explore the use of a diffusion model, trained to capture the target distribution for CT reconstruction, as an in-distribution prior. Building on recent research, we employ the model to reconstruct partially diffused input images and assess OOD-ness through multiple reconstruction errors. Adapting this approach for sparse-view CT requires redefining the notions of "input" and "reconstruction error". Here, we use filtered backprojection (FBP) reconstructions as input and investigate various definitions of reconstruction error. Our proof-of-concept experiments on the MNIST dataset highlight both successes and failures, demonstrating the potential and limitations of integrating such an OOD detector into a CT reconstruction system. Our findings suggest that effective OOD detection can be achieved by comparing measurements with forward-projected reconstructions, provided that reconstructions from noisy FBP inputs are conditioned on the measurements. However, conditioning can sometimes lead the OOD detector to inadvertently reconstruct OOD images well. To counter this, we introduce a weighting approach that improves robustness against highly informative OOD measurements, albeit with a trade-off in performance in certain cases.
Learned denoising with simulated and experimental low-dose CT data
Aug 15, 2024



Abstract:Like in many other research fields, recent developments in computational imaging have focused on developing machine learning (ML) approaches to tackle its main challenges. To improve the performance of computational imaging algorithms, machine learning methods are used for image processing tasks such as noise reduction. Generally, these ML methods heavily rely on the availability of high-quality data on which they are trained. This work explores the application of ML methods, specifically convolutional neural networks (CNNs), in the context of noise reduction for computed tomography (CT) imaging. We utilize a large 2D computed tomography dataset for machine learning to carry out for the first time a comprehensive study on the differences between the observed performances of algorithms trained on simulated noisy data and on real-world experimental noisy data. The study compares the performance of two common CNN architectures, U-Net and MSD-Net, that are trained and evaluated on both simulated and experimental noisy data. The results show that while sinogram denoising performed better with simulated noisy data if evaluated in the sinogram domain, the performance did not carry over to the reconstruction domain where training on experimental noisy data shows a higher performance in denoising experimental noisy data. Training the algorithms in an end-to-end fashion from sinogram to reconstruction significantly improved model performance, emphasizing the importance of matching raw measurement data to high-quality CT reconstructions. The study furthermore suggests the need for more sophisticated noise simulation approaches to bridge the gap between simulated and real-world data in CT image denoising applications and gives insights into the challenges and opportunities in leveraging simulated data for machine learning in computational imaging.
Experimental Validation of Ultrasound Beamforming with End-to-End Deep Learning for Single Plane Wave Imaging
Apr 22, 2024



Abstract:Ultrafast ultrasound imaging insonifies a medium with one or a combination of a few plane waves at different beam-steered angles instead of many focused waves. It can achieve much higher frame rates, but often at the cost of reduced image quality. Deep learning approaches have been proposed to mitigate this disadvantage, in particular for single plane wave imaging. Predominantly, image-to-image post-processing networks or fully learned data-to-image neural networks are used. Both construct their mapping purely data-driven and require expressive networks and large amounts of training data to perform well. In contrast, we consider data-to-image networks which incorporate a conventional image formation techniques as differentiable layers in the network architecture. This allows for end-to-end training with small amounts of training data. In this work, using f-k migration as an image formation layer is evaluated in-depth with experimental data. We acquired a data collection designed for benchmarking data-driven plane wave imaging approaches using a realistic breast mimicking phantom and an ultrasound calibration phantom. The evaluation considers global and local image similarity measures and contrast, resolution and lesion detectability analysis. The results show that the proposed network architecture is capable of improving the image quality of single plane wave images on all evaluation metrics. Furthermore, these image quality improvements can be achieved with surprisingly little amounts of training data.
Single-shot Tomography of Discrete Dynamic Objects
Nov 09, 2023



Abstract:This paper presents a novel method for the reconstruction of high-resolution temporal images in dynamic tomographic imaging, particularly for discrete objects with smooth boundaries that vary over time. Addressing the challenge of limited measurements per time point, we propose a technique that synergistically incorporates spatial and temporal information of the dynamic objects. This is achieved through the application of the level-set method for image segmentation and the representation of motion via a sinusoidal basis. The result is a computationally efficient and easily optimizable variational framework that enables the reconstruction of high-quality 2D or 3D image sequences with a single projection per frame. Compared to current methods, our proposed approach demonstrates superior performance on both synthetic and pseudo-dynamic real X-ray tomography datasets. The implications of this research extend to improved visualization and analysis of dynamic processes in tomographic imaging, finding potential applications in diverse scientific and industrial domains.
Sequential Experimental Design for X-Ray CT Using Deep Reinforcement Learning
Jul 12, 2023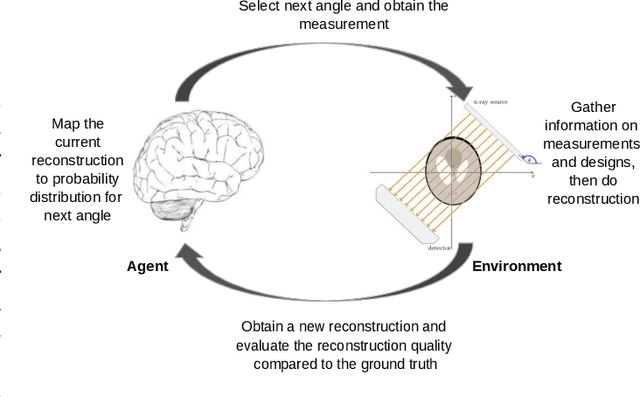
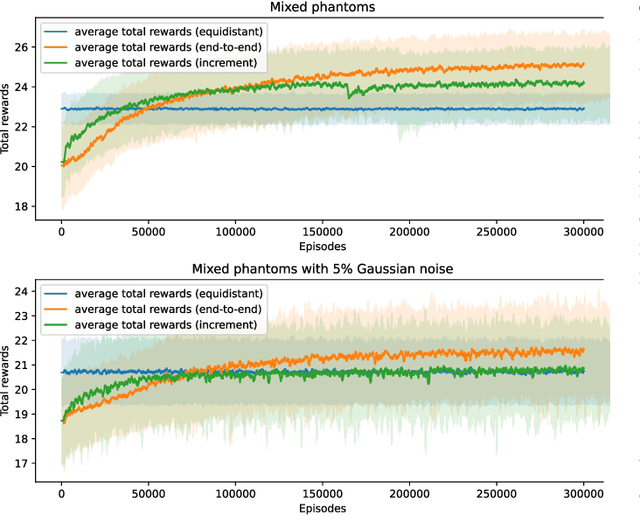
Abstract:In X-ray Computed Tomography (CT), projections from many angles are acquired and used for 3D reconstruction. To make CT suitable for in-line quality control, reducing the number of angles while maintaining reconstruction quality is necessary. Sparse-angle tomography is a popular approach for obtaining 3D reconstructions from limited data. To optimize its performance, one can adapt scan angles sequentially to select the most informative angles for each scanned object. Mathematically, this corresponds to solving and optimal experimental design (OED) problem. OED problems are high-dimensional, non-convex, bi-level optimization problems that cannot be solved online, i.e., during the scan. To address these challenges, we pose the OED problem as a partially observable Markov decision process in a Bayesian framework, and solve it through deep reinforcement learning. The approach learns efficient non-greedy policies to solve a given class of OED problems through extensive offline training rather than solving a given OED problem directly via numerical optimization. As such, the trained policy can successfully find the most informative scan angles online. We use a policy training method based on the Actor-Critic approach and evaluate its performance on 2D tomography with synthetic data.
2DeteCT -- A large 2D expandable, trainable, experimental Computed Tomography dataset for machine learning
Jun 09, 2023Abstract:Recent research in computational imaging largely focuses on developing machine learning (ML) techniques for image reconstruction, which requires large-scale training datasets consisting of measurement data and ground-truth images. However, suitable experimental datasets for X-ray Computed Tomography (CT) are scarce, and methods are often developed and evaluated only on simulated data. We fill this gap by providing the community with a versatile, open 2D fan-beam CT dataset suitable for developing ML techniques for a range of image reconstruction tasks. To acquire it, we designed a sophisticated, semi-automatic scan procedure that utilizes a highly-flexible laboratory X-ray CT setup. A diverse mix of samples with high natural variability in shape and density was scanned slice-by-slice (5000 slices in total) with high angular and spatial resolution and three different beam characteristics: A high-fidelity, a low-dose and a beam-hardening-inflicted mode. In addition, 750 out-of-distribution slices were scanned with sample and beam variations to accommodate robustness and segmentation tasks. We provide raw projection data, reference reconstructions and segmentations based on an open-source data processing pipeline.
SparseAlign: A Super-Resolution Algorithm for Automatic Marker Localization and Deformation Estimation in Cryo-Electron Tomography
Jan 21, 2022



Abstract:Tilt-series alignment is crucial to obtaining high-resolution reconstructions in cryo-electron tomography. Beam-induced local deformation of the sample is hard to estimate from the low-contrast sample alone, and often requires fiducial gold bead markers. The state-of-the-art approach for deformation estimation uses (semi-)manually labelled marker locations in projection data to fit the parameters of a polynomial deformation model. Manually-labelled marker locations are difficult to obtain when data are noisy or markers overlap in projection data. We propose an alternative mathematical approach for simultaneous marker localization and deformation estimation by extending a grid-free super-resolution algorithm first proposed in the context of single-molecule localization microscopy. Our approach does not require labelled marker locations; instead, we use an image-based loss where we compare the forward projection of markers with the observed data. We equip this marker localization scheme with an additional deformation estimation component and solve for a reduced number of deformation parameters. Using extensive numerical studies on marker-only samples, we show that our approach automatically finds markers and reliably estimates sample deformation without labelled marker data. We further demonstrate the applicability of our approach for a broad range of model mismatch scenarios, including experimental electron tomography data of gold markers on ice.
Single Plane-Wave Imaging using Physics-Based Deep Learning
Sep 08, 2021


Abstract:In plane-wave imaging, multiple unfocused ultrasound waves are transmitted into a medium of interest from different angles and an image is formed from the recorded reflections. The number of plane waves used leads to a trade-off between frame-rate and image quality, with single-plane-wave (SPW) imaging being the fastest possible modality with the worst image quality. Recently, deep learning methods have been proposed to improve ultrasound imaging. One approach is to use image-to-image networks that work on the formed image and another is to directly learn a mapping from data to an image. Both approaches utilize purely data-driven models and require deep, expressive network architectures, combined with large numbers of training samples to obtain good results. Here, we propose a data-to-image architecture that incorporates a wave-physics-based image formation algorithm in-between deep convolutional neural networks. To achieve this, we implement the Fourier (FK) migration method as network layers and train the whole network end-to-end. We compare our proposed data-to-image network with an image-to-image network in simulated data experiments, mimicking a medical ultrasound application. Experiments show that it is possible to obtain high-quality SPW images, almost similar to an image formed using 75 plane waves over an angular range of $\pm$16$^\circ$. This illustrates the great potential of combining deep neural networks with physics-based image formation algorithms for SPW imaging.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge