Poulami Somanya Ganguly
SparseAlign: A Super-Resolution Algorithm for Automatic Marker Localization and Deformation Estimation in Cryo-Electron Tomography
Jan 21, 2022



Abstract:Tilt-series alignment is crucial to obtaining high-resolution reconstructions in cryo-electron tomography. Beam-induced local deformation of the sample is hard to estimate from the low-contrast sample alone, and often requires fiducial gold bead markers. The state-of-the-art approach for deformation estimation uses (semi-)manually labelled marker locations in projection data to fit the parameters of a polynomial deformation model. Manually-labelled marker locations are difficult to obtain when data are noisy or markers overlap in projection data. We propose an alternative mathematical approach for simultaneous marker localization and deformation estimation by extending a grid-free super-resolution algorithm first proposed in the context of single-molecule localization microscopy. Our approach does not require labelled marker locations; instead, we use an image-based loss where we compare the forward projection of markers with the observed data. We equip this marker localization scheme with an additional deformation estimation component and solve for a reduced number of deformation parameters. Using extensive numerical studies on marker-only samples, we show that our approach automatically finds markers and reliably estimates sample deformation without labelled marker data. We further demonstrate the applicability of our approach for a broad range of model mismatch scenarios, including experimental electron tomography data of gold markers on ice.
Improving reproducibility in synchrotron tomography using implementation-adapted filters
Mar 15, 2021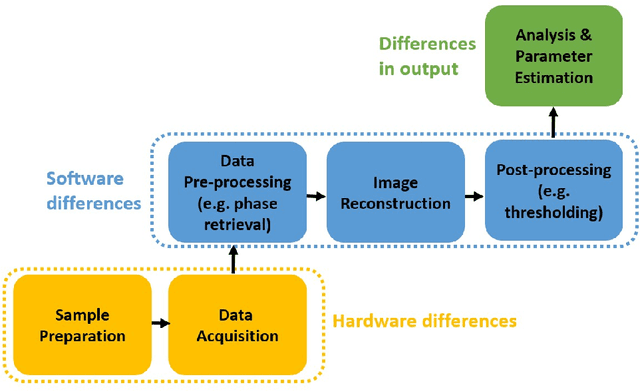

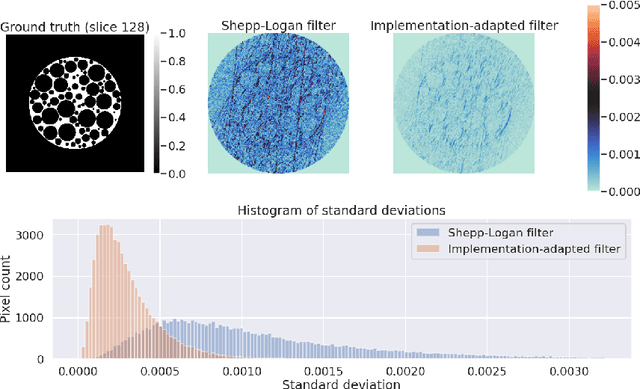

Abstract:For reconstructing large tomographic datasets fast, filtered backprojection-type or Fourier-based algorithms are still the method of choice, as they have been for decades. These robust and computationally efficient algorithms have been integrated in a broad range of software packages. Despite the fact that the underlying mathematical formulas used for image reconstruction are unambiguous, variations in discretisation and interpolation result in quantitative differences between reconstructed images obtained from different software. This hinders reproducibility of experimental results. In this paper, we propose a way to reduce such differences by optimising the filter used in analytical algorithms. These filters can be computed using a wrapper routine around a black-box implementation of a reconstruction algorithm, and lead to quantitatively similar reconstructions. We demonstrate use cases for our approach by computing implementation-adapted filters for several open-source implementations and applying it to simulated phantoms and real-world data acquired at the synchrotron. Our contribution to a reproducible reconstruction step forms a building block towards a fully reproducible synchrotron tomography data processing pipeline.
Parallel-beam X-ray CT datasets of apples with internal defects and label balancing for machine learning
Dec 24, 2020



Abstract:We present three parallel-beam tomographic datasets of 94 apples with internal defects along with defect label files. The datasets are prepared for development and testing of data-driven, learning-based image reconstruction, segmentation and post-processing methods. The three versions are a noiseless simulation; simulation with added Gaussian noise, and with scattering noise. The datasets are based on real 3D X-ray CT data and their subsequent volume reconstructions. The ground truth images, based on the volume reconstructions, are also available through this project. Apples contain various defects, which naturally introduce a label bias. We tackle this by formulating the bias as an optimization problem. In addition, we demonstrate solving this problem with two methods: a simple heuristic algorithm and through mixed integer quadratic programming. This ensures the datasets can be split into test, training or validation subsets with the label bias eliminated. Therefore the datasets can be used for image reconstruction, segmentation, automatic defect detection, and testing the effects of (as well as applying new methodologies for removing) label bias in machine learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge