Mireia Crispin-Ortuzar
A Systematic Review on Data-Driven Brain Deformation Modeling for Image-Guided Neurosurgery
Feb 09, 2026Abstract:Accurate compensation of brain deformation is a critical challenge for reliable image-guided neurosurgery, as surgical manipulation and tumor resection induce tissue motion that misaligns preoperative planning images with intraoperative anatomy and longitudinal studies. In this systematic review, we synthesize recent AI-driven approaches developed between January 2020 and April 2025 for modeling and correcting brain deformation. A comprehensive literature search was conducted in PubMed, IEEE Xplore, Scopus, and Web of Science, with predefined inclusion and exclusion criteria focused on computational methods applied to brain deformation compensation for neurosurgical imaging, resulting in 41 studies meeting these criteria. We provide a unified analysis of methodological strategies, including deep learning-based image registration, direct deformation field regression, synthesis-driven multimodal alignment, resection-aware architectures addressing missing correspondences, and hybrid models that integrate biomechanical priors. We also examine dataset utilization, reported evaluation metrics, validation protocols, and how uncertainty and generalization have been assessed across studies. While AI-based deformation models demonstrate promising performance and computational efficiency, current approaches exhibit limitations in out-of-distribution robustness, standardized benchmarking, interpretability, and readiness for clinical deployment. Our review highlights these gaps and outlines opportunities for future research aimed at achieving more robust, generalizable, and clinically translatable deformation compensation solutions for neurosurgical guidance. By organizing recent advances and critically evaluating evaluation practices, this work provides a comprehensive foundation for researchers and clinicians engaged in developing and applying AI-based brain deformation methods.
Developing Predictive and Robust Radiomics Models for Chemotherapy Response in High-Grade Serous Ovarian Carcinoma
Jan 13, 2026Abstract:Objectives: High-grade serous ovarian carcinoma (HGSOC) is typically diagnosed at an advanced stage with extensive peritoneal metastases, making treatment challenging. Neoadjuvant chemotherapy (NACT) is often used to reduce tumor burden before surgery, but about 40% of patients show limited response. Radiomics, combined with machine learning (ML), offers a promising non-invasive method for predicting NACT response by analyzing computed tomography (CT) imaging data. This study aimed to improve response prediction in HGSOC patients undergoing NACT by integration different feature selection methods. Materials and methods: A framework for selecting robust radiomics features was introduced by employing an automated randomisation algorithm to mimic inter-observer variability, ensuring a balance between feature robustness and prediction accuracy. Four response metrics were used: chemotherapy response score (CRS), RECIST, volume reduction (VolR), and diameter reduction (DiaR). Lesions in different anatomical sites were studied. Pre- and post-NACT CT scans were used for feature extraction and model training on one cohort, and an independent cohort was used for external testing. Results: The best prediction performance was achieved using all lesions combined for VolR prediction, with an AUC of 0.83. Omental lesions provided the best results for CRS prediction (AUC 0.77), while pelvic lesions performed best for DiaR (AUC 0.76). Conclusion: The integration of robustness into the feature selection processes ensures the development of reliable models and thus facilitates the implementation of the radiomics models in clinical applications for HGSOC patients. Future work should explore further applications of radiomics in ovarian cancer, particularly in real-time clinical settings.
R$^{2}$Seg: Training-Free OOD Medical Tumor Segmentation via Anatomical Reasoning and Statistical Rejection
Nov 16, 2025Abstract:Foundation models for medical image segmentation struggle under out-of-distribution (OOD) shifts, often producing fragmented false positives on OOD tumors. We introduce R$^{2}$Seg, a training-free framework for robust OOD tumor segmentation that operates via a two-stage Reason-and-Reject process. First, the Reason step employs an LLM-guided anatomical reasoning planner to localize organ anchors and generate multi-scale ROIs. Second, the Reject step applies two-sample statistical testing to candidates generated by a frozen foundation model (BiomedParse) within these ROIs. This statistical rejection filter retains only candidates significantly different from normal tissue, effectively suppressing false positives. Our framework requires no parameter updates, making it compatible with zero-update test-time augmentation and avoiding catastrophic forgetting. On multi-center and multi-modal tumor segmentation benchmarks, R$^{2}$Seg substantially improves Dice, specificity, and sensitivity over strong baselines and the original foundation models. Code are available at https://github.com/Eurekashen/R2Seg.
Integrating Pathology and CT Imaging for Personalized Recurrence Risk Prediction in Renal Cancer
Aug 29, 2025Abstract:Recurrence risk estimation in clear cell renal cell carcinoma (ccRCC) is essential for guiding postoperative surveillance and treatment. The Leibovich score remains widely used for stratifying distant recurrence risk but offers limited patient-level resolution and excludes imaging information. This study evaluates multimodal recurrence prediction by integrating preoperative computed tomography (CT) and postoperative histopathology whole-slide images (WSIs). A modular deep learning framework with pretrained encoders and Cox-based survival modeling was tested across unimodal, late fusion, and intermediate fusion setups. In a real-world ccRCC cohort, WSI-based models consistently outperformed CT-only models, underscoring the prognostic strength of pathology. Intermediate fusion further improved performance, with the best model (TITAN-CONCH with ResNet-18) approaching the adjusted Leibovich score. Random tie-breaking narrowed the gap between the clinical baseline and learned models, suggesting discretization may overstate individualized performance. Using simple embedding concatenation, radiology added value primarily through fusion. These findings demonstrate the feasibility of foundation model-based multimodal integration for personalized ccRCC risk prediction. Future work should explore more expressive fusion strategies, larger multimodal datasets, and general-purpose CT encoders to better match pathology modeling capacity.
ST-Prompt Guided Histological Hypergraph Learning for Spatial Gene Expression Prediction
Mar 21, 2025



Abstract:Spatial Transcriptomics (ST) reveals the spatial distribution of gene expression in tissues, offering critical insights into biological processes and disease mechanisms. However, predicting ST from H\&E-stained histology images is challenging due to the heterogeneous relationship between histomorphology and gene expression, which arises from substantial variability across different patients and tissue sections. A more practical and valuable approach is to utilize ST data from a few local regions to predict the spatial transcriptomic landscape across the remaining regions in H&E slides. In response, we propose PHG2ST, an ST-prompt guided histological hypergraph learning framework, which leverages sparse ST signals as prompts to guide histological hypergraph learning for global spatial gene expression prediction. Our framework fuses histological hypergraph representations at multiple scales through a masked ST-prompt encoding mechanism, improving robustness and generalizability. Benchmark evaluations on two public ST datasets demonstrate that PHG2ST outperforms the existing state-of-the-art methods and closely aligns with the ground truth. These results underscore the potential of leveraging sparse local ST data for scalable and cost-effective spatial gene expression mapping in real-world biomedical applications.
CTARR: A fast and robust method for identifying anatomical regions on CT images via atlas registration
Oct 03, 2024Abstract:Medical image analysis tasks often focus on regions or structures located in a particular location within the patient's body. Often large parts of the image may not be of interest for the image analysis task. When using deep-learning based approaches, this causes an unnecessary increases the computational burden during inference and raises the chance of errors. In this paper, we introduce CTARR, a novel generic method for CT Anatomical Region Recognition. The method serves as a pre-processing step for any deep learning-based CT image analysis pipeline by automatically identifying the pre-defined anatomical region that is relevant for the follow-up task and removing the rest. It can be used in (i) image segmentation to prevent false positives in anatomically implausible regions and speeding up the inference, (ii) image classification to produce image crops that are consistent in their anatomical context, and (iii) image registration by serving as a fast pre-registration step. Our proposed method is based on atlas registration and provides a fast and robust way to crop any anatomical region encoded as one or multiple bounding box(es) from any unlabeled CT scan of the brain, chest, abdomen and/or pelvis. We demonstrate the utility and robustness of the proposed method in the context of medical image segmentation by evaluating it on six datasets of public segmentation challenges. The foreground voxels in the regions of interest are preserved in the vast majority of cases and tasks (97.45-100%) while taking only fractions of a seconds to compute (0.1-0.21s) on a deep learning workstation and greatly reducing the segmentation runtime (2.0-12.7x). Our code is available at https://github.com/ThomasBudd/ctarr.
CoxKAN: Kolmogorov-Arnold Networks for Interpretable, High-Performance Survival Analysis
Sep 06, 2024Abstract:Survival analysis is a branch of statistics used for modeling the time until a specific event occurs and is widely used in medicine, engineering, finance, and many other fields. When choosing survival models, there is typically a trade-off between performance and interpretability, where the highest performance is achieved by black-box models based on deep learning. This is a major problem in fields such as medicine where practitioners are reluctant to blindly trust black-box models to make important patient decisions. Kolmogorov-Arnold Networks (KANs) were recently proposed as an interpretable and accurate alternative to multi-layer perceptrons (MLPs). We introduce CoxKAN, a Cox proportional hazards Kolmogorov-Arnold Network for interpretable, high-performance survival analysis. We evaluate the proposed CoxKAN on 4 synthetic datasets and 9 real medical datasets. The synthetic experiments demonstrate that CoxKAN accurately recovers interpretable symbolic formulae for the hazard function, and effectively performs automatic feature selection. Evaluation on the 9 real datasets show that CoxKAN consistently outperforms the Cox proportional hazards model and achieves performance that is superior or comparable to that of tuned MLPs. Furthermore, we find that CoxKAN identifies complex interactions between predictor variables that would be extremely difficult to recognise using existing survival methods, and automatically finds symbolic formulae which uncover the precise effect of important biomarkers on patient risk.
Calibrating Ensembles for Scalable Uncertainty Quantification in Deep Learning-based Medical Segmentation
Sep 20, 2022
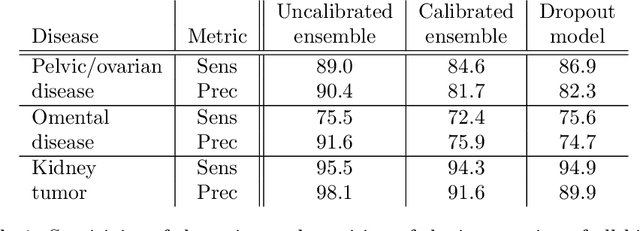


Abstract:Uncertainty quantification in automated image analysis is highly desired in many applications. Typically, machine learning models in classification or segmentation are only developed to provide binary answers; however, quantifying the uncertainty of the models can play a critical role for example in active learning or machine human interaction. Uncertainty quantification is especially difficult when using deep learning-based models, which are the state-of-the-art in many imaging applications. The current uncertainty quantification approaches do not scale well in high-dimensional real-world problems. Scalable solutions often rely on classical techniques, such as dropout, during inference or training ensembles of identical models with different random seeds to obtain a posterior distribution. In this paper, we show that these approaches fail to approximate the classification probability. On the contrary, we propose a scalable and intuitive framework to calibrate ensembles of deep learning models to produce uncertainty quantification measurements that approximate the classification probability. On unseen test data, we demonstrate improved calibration, sensitivity (in two out of three cases) and precision when being compared with the standard approaches. We further motivate the usage of our method in active learning, creating pseudo-labels to learn from unlabeled images and human-machine collaboration.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge