Isabella Ellinger
Improving Generalization Capability of Deep Learning-Based Nuclei Instance Segmentation by Non-deterministic Train Time and Deterministic Test Time Stain Normalization
Sep 12, 2023



Abstract:With the advent of digital pathology and microscopic systems that can scan and save whole slide histological images automatically, there is a growing trend to use computerized methods to analyze acquired images. Among different histopathological image analysis tasks, nuclei instance segmentation plays a fundamental role in a wide range of clinical and research applications. While many semi- and fully-automatic computerized methods have been proposed for nuclei instance segmentation, deep learning (DL)-based approaches have been shown to deliver the best performances. However, the performance of such approaches usually degrades when tested on unseen datasets. In this work, we propose a novel approach to improve the generalization capability of a DL-based automatic segmentation approach. Besides utilizing one of the state-of-the-art DL-based models as a baseline, our method incorporates non-deterministic train time and deterministic test time stain normalization. We trained the model with one single training set and evaluated its segmentation performance on seven test datasets. Our results show that the proposed method provides up to 5.77%, 5.36%, and 5.27% better performance in segmenting nuclei based on Dice score, aggregated Jaccard index, and panoptic quality score, respectively, compared to the baseline segmentation model.
NuInsSeg: A Fully Annotated Dataset for Nuclei Instance Segmentation in H&E-Stained Histological Images
Aug 03, 2023Abstract:In computational pathology, automatic nuclei instance segmentation plays an essential role in whole slide image analysis. While many computerized approaches have been proposed for this task, supervised deep learning (DL) methods have shown superior segmentation performances compared to classical machine learning and image processing techniques. However, these models need fully annotated datasets for training which is challenging to acquire, especially in the medical domain. In this work, we release one of the biggest fully manually annotated datasets of nuclei in Hematoxylin and Eosin (H&E)-stained histological images, called NuInsSeg. This dataset contains 665 image patches with more than 30,000 manually segmented nuclei from 31 human and mouse organs. Moreover, for the first time, we provide additional ambiguous area masks for the entire dataset. These vague areas represent the parts of the images where precise and deterministic manual annotations are impossible, even for human experts. The dataset and detailed step-by-step instructions to generate related segmentation masks are publicly available at https://www.kaggle.com/datasets/ipateam/nuinsseg and https://github.com/masih4/NuInsSeg, respectively.
Deep Neural Network Pruning for Nuclei Instance Segmentation in Hematoxylin & Eosin-Stained Histological Images
Jun 15, 2022
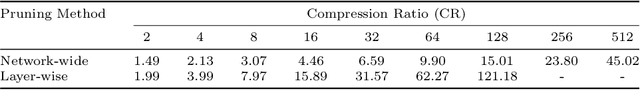

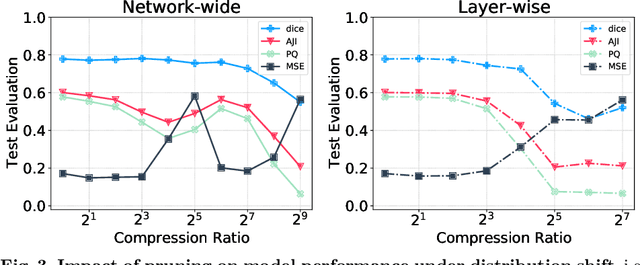
Abstract:Recently, pruning deep neural networks (DNNs) has received a lot of attention for improving accuracy and generalization power, reducing network size, and increasing inference speed on specialized hardwares. Although pruning was mainly tested on computer vision tasks, its application in the context of medical image analysis has hardly been explored. This work investigates the impact of well-known pruning techniques, namely layer-wise and network-wide magnitude pruning, on the nuclei instance segmentation performance in histological images. Our utilized instance segmentation model consists of two main branches: (1) a semantic segmentation branch, and (2) a deep regression branch. We investigate the impact of weight pruning on the performance of both branches separately and on the final nuclei instance segmentation result. Evaluated on two publicly available datasets, our results show that layer-wise pruning delivers slightly better performance than networkwide pruning for small compression ratios (CRs) while for large CRs, network-wide pruning yields superior performance. For semantic segmentation, deep regression and final instance segmentation, 93.75 %, 95 %, and 80 % of the model weights can be pruned by layer-wise pruning with less than 2 % reduction in the performance of respective models.
FUSeg: The Foot Ulcer Segmentation Challenge
Jan 02, 2022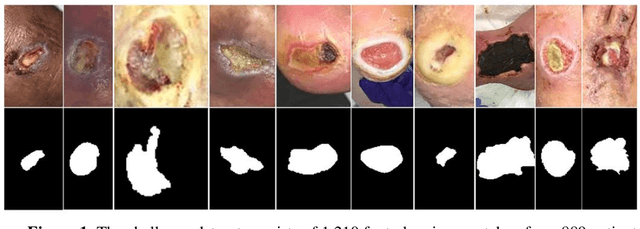
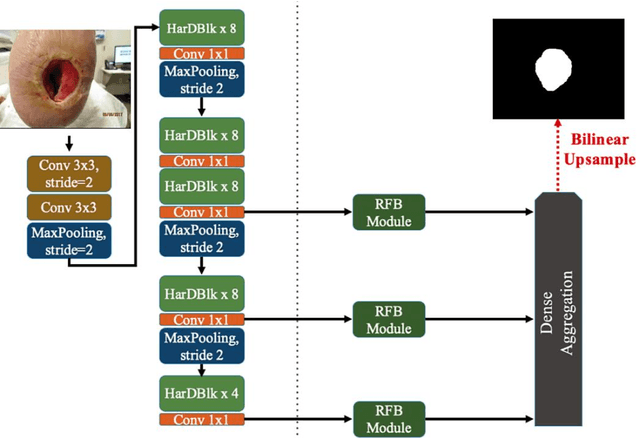
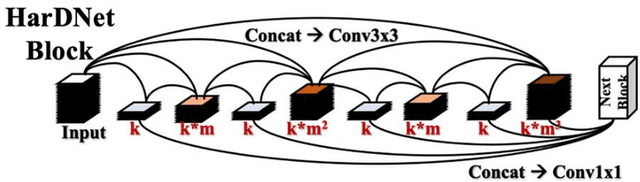
Abstract:Acute and chronic wounds with varying etiologies burden the healthcare systems economically. The advanced wound care market is estimated to reach $22 billion by 2024. Wound care professionals provide proper diagnosis and treatment with heavy reliance on images and image documentation. Segmentation of wound boundaries in images is a key component of the care and diagnosis protocol since it is important to estimate the area of the wound and provide quantitative measurement for the treatment. Unfortunately, this process is very time-consuming and requires a high level of expertise. Recently automatic wound segmentation methods based on deep learning have shown promising performance but require large datasets for training and it is unclear which methods perform better. To address these issues, we propose the Foot Ulcer Segmentation challenge (FUSeg) organized in conjunction with the 2021 International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI). We built a wound image dataset containing 1,210 foot ulcer images collected over 2 years from 889 patients. It is pixel-wise annotated by wound care experts and split into a training set with 1010 images and a testing set with 200 images for evaluation. Teams around the world developed automated methods to predict wound segmentations on the testing set of which annotations were kept private. The predictions were evaluated and ranked based on the average Dice coefficient. The FUSeg challenge remains an open challenge as a benchmark for wound segmentation after the conference.
Automatic Foot Ulcer segmentation Using an Ensemble of Convolutional Neural Networks
Sep 03, 2021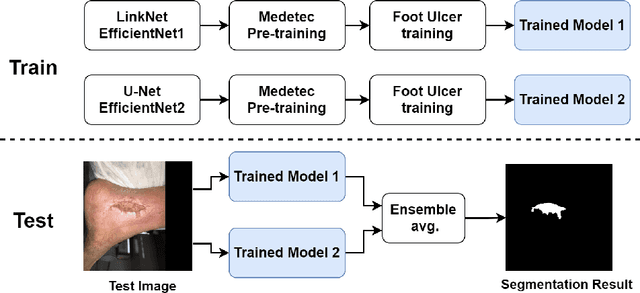
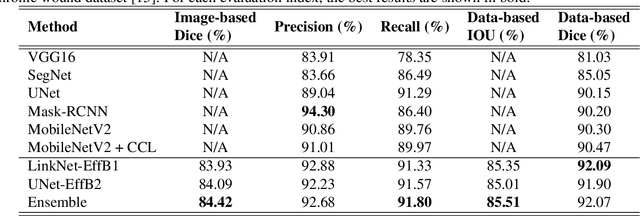
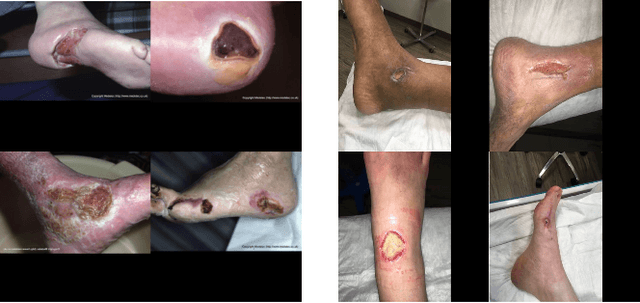

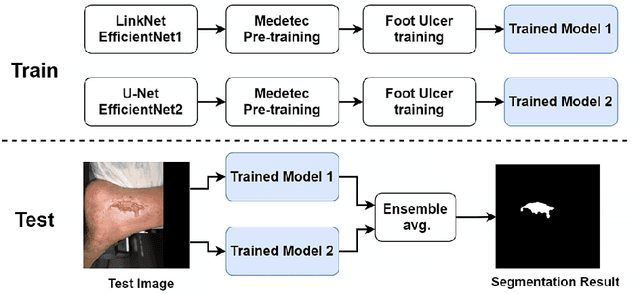
Abstract:Foot ulcer is a common complication of diabetes mellitus; it is associated with substantial morbidity and mortality and remains a major risk factor for lower leg amputation. Extracting accurate morphological features from the foot wounds is crucial for proper treatment. Although visual and manual inspection by medical professionals is the common approach to extract the features, this method is subjective and error-prone. Computer-mediated approaches are the alternative solutions to segment the lesions and extract related morphological features. Among various proposed computer-based approaches for image segmentation, deep learning-based methods and more specifically convolutional neural networks (CNN) have shown excellent performances for various image segmentation tasks including medical image segmentation. In this work, we proposed an ensemble approach based on two encoder-decoder-based CNN models, namely LinkNet and UNet, to perform foot ulcer segmentation. To deal with limited training samples, we used pre-trained weights (EfficientNetB1 for the LinkNet model and EfficientNetB2 for the UNet model) and further pre-training by the Medetec dataset. We also applied a number of morphological-based and colour-based augmentation techniques to train the models. We integrated five-fold cross-validation, test time augmentation and result fusion in our proposed ensemble approach to boost the segmentation performance. Applied on a publicly available foot ulcer segmentation dataset and the MICCAI 2021 Foot Ulcer Segmentation (FUSeg) Challenge, our method achieved state-of-the-art data-based Dice scores of 92.07% and 88.80%, respectively. Our developed method achieved the first rank in the FUSeg challenge leaderboard. The Dockerised guideline, inference codes and saved trained models are publicly available in the published GitHub repository: https://github.com/masih4/Foot_Ulcer_Segmentation
CryoNuSeg: A Dataset for Nuclei Instance Segmentation of Cryosectioned H&E-Stained Histological Images
Jan 02, 2021
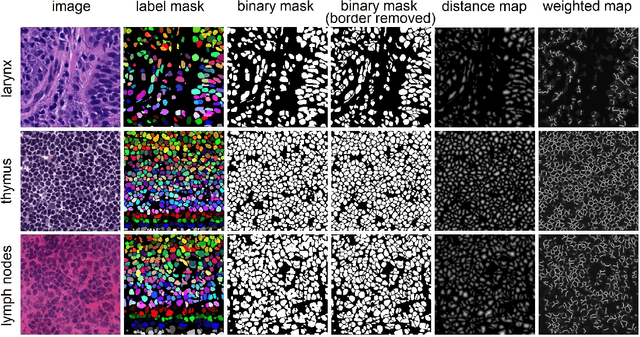
Abstract:Nuclei instance segmentation plays an important role in the analysis of Hematoxylin and Eosin (H&E)-stained images. While supervised deep learning (DL)-based approaches represent the state-of-the-art in automatic nuclei instance segmentation, annotated datasets are required to train these models. There are two main types of tissue processing protocols, namely formalin-fixed paraffin-embedded samples (FFPE) and frozen tissue samples (FS). Although FFPE-derived H&E stained tissue sections are the most widely used samples, H&E staining on frozen sections derived from FS samples is a relevant method in intra-operative surgical sessions as it can be performed fast. Due to differences in the protocols of these two types of samples, the derived images and in particular the nuclei appearance may be different in the acquired whole slide images. Analysis of FS-derived H&E stained images can be more challenging as rapid preparation, staining, and scanning of FS sections may lead to deterioration in image quality. In this paper, we introduce CryoNuSeg, the first fully annotated FS-derived cryosectioned and H&E-stained nuclei instance segmentation dataset. The dataset contains images from 10 human organs that were not exploited in other publicly available datasets, and is provided with three manual mark-ups to allow measuring intra-observer and inter-observer variability. Moreover, we investigate the effects of tissue fixation/embedding protocol (i.e., FS or FFPE) on the automatic nuclei instance segmentation performance of one of the state-of-the-art DL approaches. We also create a baseline segmentation benchmark for the dataset that can be used in future research. A step-by-step guide to generate the dataset as well as the full dataset and other detailed information are made available to fellow researchers at https://github.com/masih4/CryoNuSeg.
Pollen Grain Microscopic Image Classification Using an Ensemble of Fine-Tuned Deep Convolutional Neural Networks
Nov 15, 2020


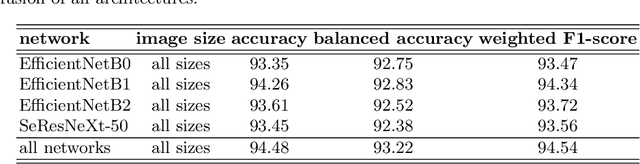
Abstract:Pollen grain micrograph classification has multiple applications in medicine and biology. Automatic pollen grain image classification can alleviate the problems of manual categorisation such as subjectivity and time constraints. While a number of computer-based methods have been introduced in the literature to perform this task, classification performance needs to be improved for these methods to be useful in practice. In this paper, we present an ensemble approach for pollen grain microscopic image classification into four categories: Corylus Avellana well-developed pollen grain, Corylus Avellana anomalous pollen grain, Alnus well-developed pollen grain, and non-pollen (debris) instances. In our approach, we develop a classification strategy that is based on fusion of four state-of-the-art fine-tuned convolutional neural networks, namely EfficientNetB0, EfficientNetB1, EfficientNetB2 and SeResNeXt-50 deep models. These models are trained with images of three fixed sizes (224x224, 240x240, and 260x260 pixels) and their prediction probability vectors are then fused in an ensemble method to form a final classification vector for a given pollen grain image. Our proposed method is shown to yield excellent classification performance, obtaining an accuracy of of 94.48% and a weighted F1-score of 94.54% on the ICPR 2020 Pollen Grain Classification Challenge training dataset based on five-fold cross-validation. Evaluated on the test set of the challenge, our approach achieved a very competitive performance in comparison to the top ranked approaches with an accuracy and a weighted F1-score of 96.28% and 96.30%, respectively.
The Effects of Skin Lesion Segmentation on the Performance of Dermatoscopic Image Classification
Aug 28, 2020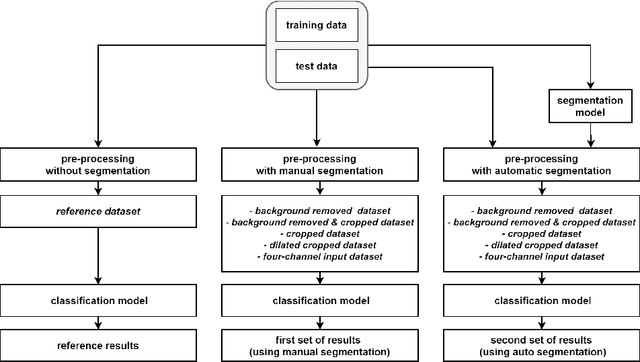
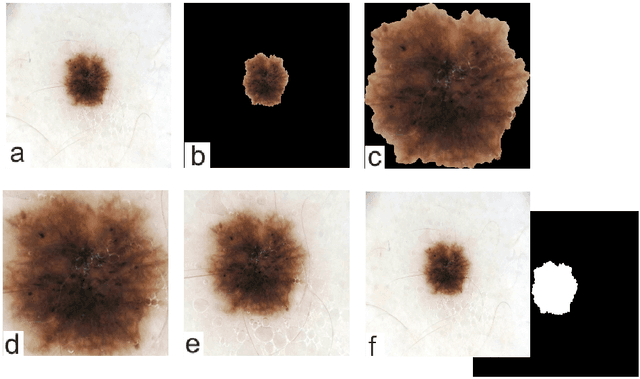
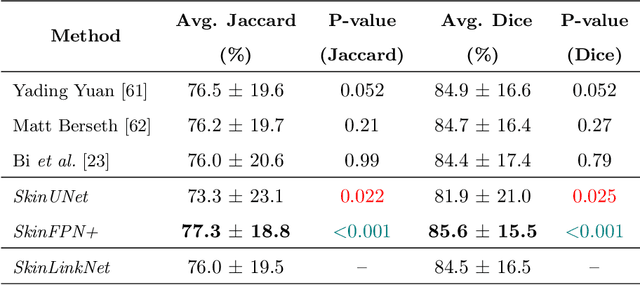
Abstract:Malignant melanoma (MM) is one of the deadliest types of skin cancer. Analysing dermatoscopic images plays an important role in the early detection of MM and other pigmented skin lesions. Among different computer-based methods, deep learning-based approaches and in particular convolutional neural networks have shown excellent classification and segmentation performances for dermatoscopic skin lesion images. These models can be trained end-to-end without requiring any hand-crafted features. However, the effect of using lesion segmentation information on classification performance has remained an open question. In this study, we explicitly investigated the impact of using skin lesion segmentation masks on the performance of dermatoscopic image classification. To do this, first, we developed a baseline classifier as the reference model without using any segmentation masks. Then, we used either manually or automatically created segmentation masks in both training and test phases in different scenarios and investigated the classification performances. Evaluated on the ISIC 2017 challenge dataset which contained two binary classification tasks (i.e. MM vs. all and seborrheic keratosis (SK) vs. all) and based on the derived area under the receiver operating characteristic curve scores, we observed four main outcomes. Our results show that 1) using segmentation masks did not significantly improve the MM classification performance in any scenario, 2) in one of the scenarios (using segmentation masks for dilated cropping), SK classification performance was significantly improved, 3) removing all background information by the segmentation masks significantly degraded the overall classification performance, and 4) in case of using the appropriate scenario (using segmentation for dilated cropping), there is no significant difference of using manually or automatically created segmentation masks.
Investigating and Exploiting Image Resolution for Transfer Learning-based Skin Lesion Classification
Jun 25, 2020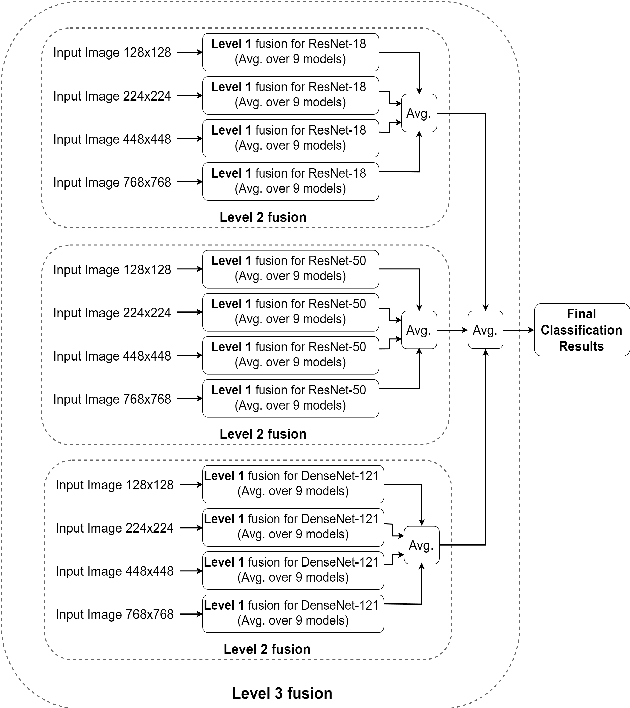

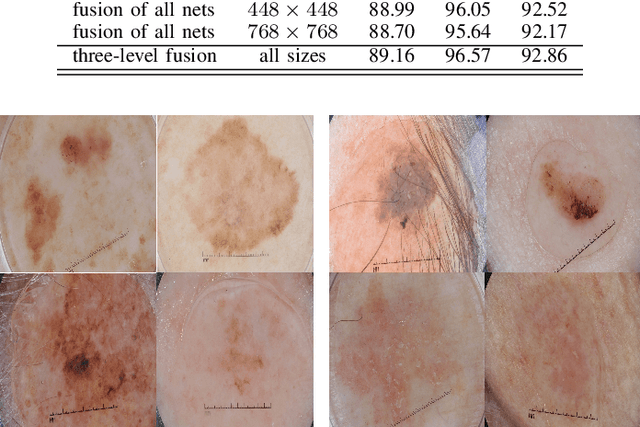

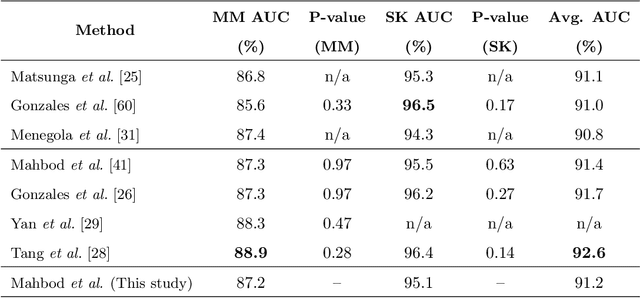
Abstract:Skin cancer is among the most common cancer types. Dermoscopic image analysis improves the diagnostic accuracy for detection of malignant melanoma and other pigmented skin lesions when compared to unaided visual inspection. Hence, computer-based methods to support medical experts in the diagnostic procedure are of great interest. Fine-tuning pre-trained convolutional neural networks (CNNs) has been shown to work well for skin lesion classification. Pre-trained CNNs are usually trained with natural images of a fixed image size which is typically significantly smaller than captured skin lesion images and consequently dermoscopic images are downsampled for fine-tuning. However, useful medical information may be lost during this transformation. In this paper, we explore the effect of input image size on skin lesion classification performance of fine-tuned CNNs. For this, we resize dermoscopic images to different resolutions, ranging from 64x64 to 768x768 pixels and investigate the resulting classification performance of three well-established CNNs, namely DenseNet-121, ResNet-18, and ResNet-50. Our results show that using very small images (of size 64x64 pixels) degrades the classification performance, while images of size 128x128 pixels and above support good performance with larger image sizes leading to slightly improved classification. We further propose a novel fusion approach based on a three-level ensemble strategy that exploits multiple fine-tuned networks trained with dermoscopic images at various sizes. When applied on the ISIC 2017 skin lesion classification challenge, our fusion approach yields an area under the receiver operating characteristic curve of 89.2% and 96.6% for melanoma classification and seborrheic keratosis classification, respectively, outperforming state-of-the-art algorithms.
Skin Lesion Classification Using Hybrid Deep Neural Networks
Feb 27, 2017
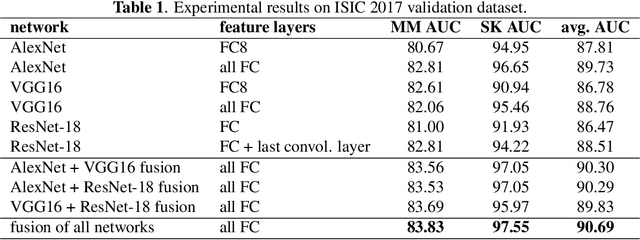
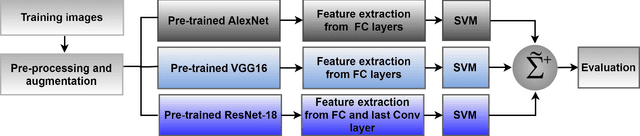
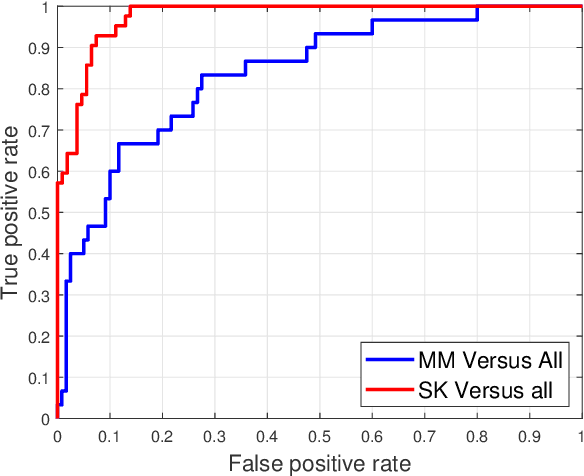
Abstract:Skin cancer is one of the major types of cancers and its incidence has been increasing over the past decades. Skin lesions can arise from various dermatologic disorders and can be classified to various types according to their texture, structure, color and other morphological features. The accuracy of diagnosis of skin lesions, specifically the discrimination of benign and malignant lesions, is paramount to ensure appropriate patient treatment. Machine learning-based classification approaches are among popular automatic methods for skin lesion classification. While there are many existing methods, convolutional neural networks (CNN) have shown to be superior over other classical machine learning methods for object detection and classification tasks. In this work, a fully automatic computerized method is proposed, which employs well established pre-trained convolutional neural networks and ensembles learning to classify skin lesions. We trained the networks using 2000 skin lesion images available from the ISIC 2017 challenge, which has three main categories and includes 374 melanoma, 254 seborrheic keratosis and 1372 benign nevi images. The trained classifier was then tested on 150 unlabeled images. The results, evaluated by the challenge organizer and based on the area under the receiver operating characteristic curve (AUC), were 84.8% and 93.6% for Melanoma and seborrheic keratosis binary classification problem, respectively. The proposed method achieved competitive results to experienced dermatologist. Further improvement and optimization of the proposed method with a larger training dataset could lead to a more precise, reliable and robust method for skin lesion classification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge