CryoNuSeg: A Dataset for Nuclei Instance Segmentation of Cryosectioned H&E-Stained Histological Images
Paper and Code
Jan 02, 2021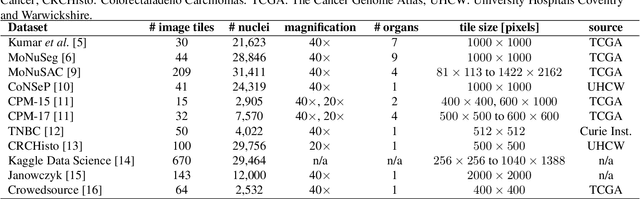
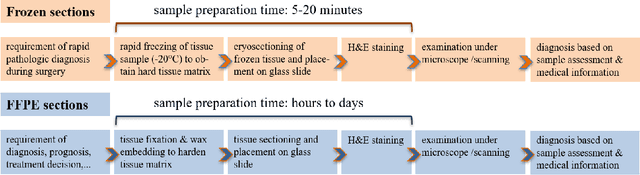
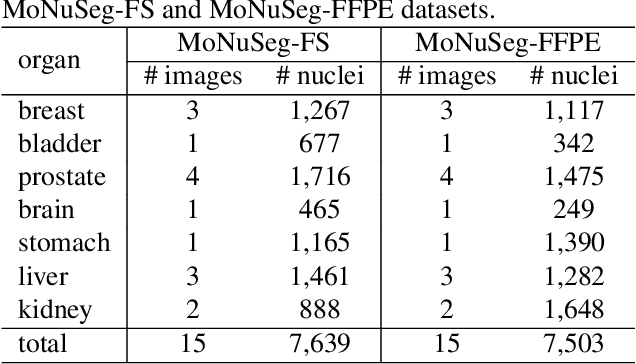
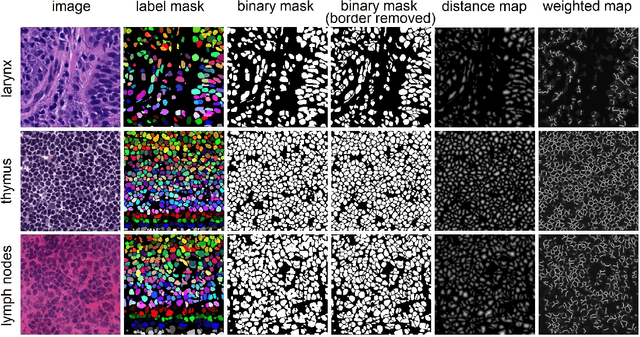
Nuclei instance segmentation plays an important role in the analysis of Hematoxylin and Eosin (H&E)-stained images. While supervised deep learning (DL)-based approaches represent the state-of-the-art in automatic nuclei instance segmentation, annotated datasets are required to train these models. There are two main types of tissue processing protocols, namely formalin-fixed paraffin-embedded samples (FFPE) and frozen tissue samples (FS). Although FFPE-derived H&E stained tissue sections are the most widely used samples, H&E staining on frozen sections derived from FS samples is a relevant method in intra-operative surgical sessions as it can be performed fast. Due to differences in the protocols of these two types of samples, the derived images and in particular the nuclei appearance may be different in the acquired whole slide images. Analysis of FS-derived H&E stained images can be more challenging as rapid preparation, staining, and scanning of FS sections may lead to deterioration in image quality. In this paper, we introduce CryoNuSeg, the first fully annotated FS-derived cryosectioned and H&E-stained nuclei instance segmentation dataset. The dataset contains images from 10 human organs that were not exploited in other publicly available datasets, and is provided with three manual mark-ups to allow measuring intra-observer and inter-observer variability. Moreover, we investigate the effects of tissue fixation/embedding protocol (i.e., FS or FFPE) on the automatic nuclei instance segmentation performance of one of the state-of-the-art DL approaches. We also create a baseline segmentation benchmark for the dataset that can be used in future research. A step-by-step guide to generate the dataset as well as the full dataset and other detailed information are made available to fellow researchers at https://github.com/masih4/CryoNuSeg.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge